Defining the Essential Function of Yeast Hsf1 Reveals a Compact Transcriptional Program for Maintaining Eukaryotic Proteostasis
- PMID: 27320198
- PMCID: PMC4938784
- DOI: 10.1016/j.molcel.2016.05.014
Defining the Essential Function of Yeast Hsf1 Reveals a Compact Transcriptional Program for Maintaining Eukaryotic Proteostasis
Erratum in
-
Defining the Essential Function of Yeast Hsf1 Reveals a Compact Transcriptional Program for Maintaining Eukaryotic Proteostasis.Mol Cell. 2018 Feb 1;69(3):534. doi: 10.1016/j.molcel.2018.01.021. Mol Cell. 2018. PMID: 29395071 Free PMC article. No abstract available.
Abstract
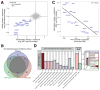
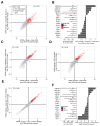
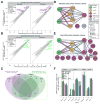
Despite its eponymous association with the heat shock response, yeast heat shock factor 1 (Hsf1) is essential even at low temperatures. Here we show that engineered nuclear export of Hsf1 results in cytotoxicity associated with massive protein aggregation. Genome-wide analysis revealed that Hsf1 nuclear export immediately decreased basal transcription and mRNA expression of 18 genes, which predominately encode chaperones. Strikingly, rescuing basal expression of Hsp70 and Hsp90 chaperones enabled robust cell growth in the complete absence of Hsf1. With the exception of chaperone gene induction, the vast majority of the heat shock response was Hsf1 independent. By comparative analysis of mammalian cell lines, we found that only heat shock-induced but not basal expression of chaperones is dependent on the mammalian Hsf1 homolog (HSF1). Our work reveals that yeast chaperone gene expression is an essential housekeeping mechanism and provides a roadmap for defining the function of HSF1 as a driver of oncogenesis.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Heat Shock Factor 1: From Fire Chief to Crowd-Control Specialist.Mol Cell. 2016 Jul 7;63(1):1-2. doi: 10.1016/j.molcel.2016.06.026. Mol Cell. 2016. PMID: 27392142
References
-
- Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. - PubMed
-
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. - PubMed
-
- Boy-Marcotte E, Lagniel G, Perrot M, Bussereau F, Boudsocq A, Jacquet M, Labarre J. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Molecular Microbiology. 1999;33:274–283. - PubMed
-
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

