Ultrasensitive proteomic quantitation of cellular signaling by digitized nanoparticle-protein counting
- PMID: 27320899
- PMCID: PMC4913309
- DOI: 10.1038/srep28163
Ultrasensitive proteomic quantitation of cellular signaling by digitized nanoparticle-protein counting
Abstract
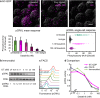
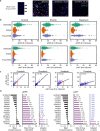
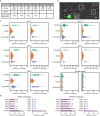
Many important signaling and regulatory proteins are expressed at low abundance and are difficult to measure in single cells. We report a molecular imaging approach to quantitate protein levels by digitized, discrete counting of nanoparticle-tagged proteins. Digitized protein counting provides ultrasensitive molecular detection of proteins in single cells that surpasses conventional methods of quantitating total diffuse fluorescence, and offers a substantial improvement in protein quantitation. We implement this digitized proteomic approach in an integrated imaging platform, the single cell-quantum dot platform (SC-QDP), to execute sensitive single cell phosphoquantitation in response to multiple drug treatment conditions and using limited primary patient material. The SC-QDP: 1) identified pAKT and pERK phospho-heterogeneity and insensitivity in individual leukemia cells treated with a multi-drug panel of FDA-approved kinase inhibitors, and 2) revealed subpopulations of drug-insensitive CD34+ stem cells with high pCRKL and pSTAT5 signaling in chronic myeloid leukemia patient blood samples. This ultrasensitive digitized protein detection approach is valuable for uncovering subtle but important differences in signaling, drug insensitivity, and other key cellular processes amongst single cells.
Figures

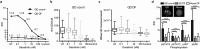
References
-
- Mann M., Hendrickson R. C. & Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu Rev Biochem 70, 437–473 (2001). - PubMed
-
- Twyman R. Principles Of Proteomics. Garland Science (2013).
-
- Paradela A. & Albar J. P. Advances in the Analysis of Protein Phosphorylation. Journal of proteome research 7, 1809–1818 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

