Mechanical Genomics Identifies Diverse Modulators of Bacterial Cell Stiffness
- PMID: 27321372
- PMCID: PMC4967499
- DOI: 10.1016/j.cels.2016.05.006
Mechanical Genomics Identifies Diverse Modulators of Bacterial Cell Stiffness
Abstract
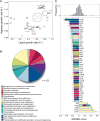
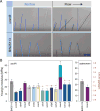
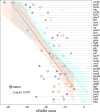
Bacteria must maintain mechanical integrity to withstand the large osmotic pressure differential across the cell membrane and wall. Although maintaining mechanical integrity is critical for proper cellular function, a fact exploited by prominent cell-wall-targeting antibiotics, the proteins that contribute to cellular mechanics remain unidentified. Here, we describe a high-throughput optical method for quantifying cell stiffness and apply this technique to a genome-wide collection of ∼4,000 Escherichia coli mutants. We identify genes with roles in diverse functional processes spanning cell-wall synthesis, energy production, and DNA replication and repair that significantly change cell stiffness when deleted. We observe that proteins with biochemically redundant roles in cell-wall synthesis exhibit different stiffness defects when deleted. Correlating our data with chemical screens reveals that reducing membrane potential generally increases cell stiffness. In total, our work demonstrates that bacterial cell stiffness is a property of both the cell wall and broader cell physiology and lays the groundwork for future systematic studies of mechanoregulation.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
-
- Bertsche U, Breukink E, Kast T, Vollmer W. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. The Journal of biological chemistry. 2005;280:38096–38101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

