Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways
- PMID: 27321668
- PMCID: PMC4987707
- DOI: 10.1039/c6np00035e
Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways
Abstract
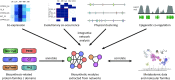
Covering: 2003 to 2016The last decade has seen the first major discoveries regarding the genomic basis of plant natural product biosynthetic pathways. Four key computationally driven strategies have been developed to identify such pathways, which make use of physical clustering, co-expression, evolutionary co-occurrence and epigenomic co-regulation of the genes involved in producing a plant natural product. Here, we discuss how these approaches can be used for the discovery of plant biosynthetic pathways encoded by both chromosomally clustered and non-clustered genes. Additionally, we will discuss opportunities to prioritize plant gene clusters for experimental characterization, and end with a forward-looking perspective on how synthetic biology technologies will allow effective functional reconstitution of candidate pathways using a variety of genetic systems.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- BB/L014130/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M028860/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/00000614/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U01 GM110699/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

