Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD
- PMID: 27321670
- PMCID: PMC4946995
- DOI: 10.1016/j.cell.2016.05.048
Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD
Abstract
Misfolded proteins of the ER are retrotranslocated to the cytosol, where they are polyubiquitinated, extracted from the membrane, and degraded by the proteasome. To investigate how the ER-associated Degradation (ERAD) machinery can accomplish retrotranslocation of a misfolded luminal protein domain across a lipid bilayer, we have reconstituted retrotranslocation with purified S. cerevisiae proteins, using proteoliposomes containing the multi-spanning ubiquitin ligase Hrd1. Retrotranslocation of the luminal domain of a membrane-spanning substrate is triggered by autoubiquitination of Hrd1. Substrate ubiquitination is a subsequent event, and the Cdc48 ATPase that completes substrate extraction from the membrane is not required for retrotranslocation. Ubiquitination of lysines in Hrd1's RING-finger domain is required for substrate retrotranslocation in vitro and for ERAD in vivo. Our results suggest that Hrd1 forms a ubiquitin-gated protein-conducting channel.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
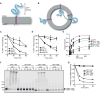




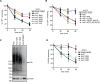

Comment in
-
Ever HRD a ubiquitin-gated channel?Cell Res. 2016 Oct;26(10):1075-1076. doi: 10.1038/cr.2016.92. Epub 2016 Aug 5. Cell Res. 2016. PMID: 27491350 Free PMC article.
References
-
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24:701–711. - PubMed
-
- Bordallo J, Wolf DH. A RING-H2 finger motif is essential for the function of Der3/Hrd1 in endoplasmic reticulum associated protein degradation in the yeast Saccharomyces cerevisiae. FEBS Lett. 1999;448:244–248. - PubMed
-
- Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

