Dynamic Axonal Translation in Developing and Mature Visual Circuits
- PMID: 27321671
- PMCID: PMC4930487
- DOI: 10.1016/j.cell.2016.05.029
Dynamic Axonal Translation in Developing and Mature Visual Circuits
Abstract
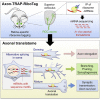
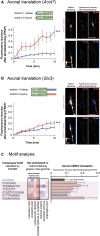
Local mRNA translation mediates the adaptive responses of axons to extrinsic signals, but direct evidence that it occurs in mammalian CNS axons in vivo is scant. We developed an axon-TRAP-RiboTag approach in mouse that allows deep-sequencing analysis of ribosome-bound mRNAs in the retinal ganglion cell axons of the developing and adult retinotectal projection in vivo. The embryonic-to-postnatal axonal translatome comprises an evolving subset of enriched genes with axon-specific roles, suggesting distinct steps in axon wiring, such as elongation, pruning, and synaptogenesis. Adult axons, remarkably, have a complex translatome with strong links to axon survival, neurotransmission, and neurodegenerative disease. Translationally co-regulated mRNA subsets share common upstream regulators, and sequence elements generated by alternative splicing promote axonal mRNA translation. Our results indicate that intricate regulation of compartment-specific mRNA translation in mammalian CNS axons supports the formation and maintenance of neural circuits in vivo.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures










References
-
- Aakalu G., Smith W.B., Nguyen N., Jiang C., Schuman E.M. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. - PubMed
-
- Andreassi C., Zimmermann C., Mitter R., Fusco S., De Vita S., Saiardi A., Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 2010;13:291–301. - PubMed
-
- Bagni C., Greenough W.T. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat. Rev. Neurosci. 2005;6:376–387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

