Role of the p55-gamma subunit of PI3K in ALK-induced cell migration: RNAi-based selection of cell migration regulators
- PMID: 27322022
- PMCID: PMC5479453
- DOI: 10.1080/19336918.2016.1202385
Role of the p55-gamma subunit of PI3K in ALK-induced cell migration: RNAi-based selection of cell migration regulators
Abstract
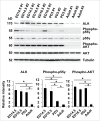
Recently, unbiased functional genetic selection identified novel cell migration-regulating genes. This RNAi-based functional selection was performed using 63,996 pooled lentiviral shRNAs targeting 21,332 mouse genes. After five rounds of selection using cells with accelerated or impaired migration, shRNAs were retrieved and identified by half-hairpin barcode sequencing using cells with the selected phenotypes. This selection process led to the identification of 29 novel cell migration regulators. One of these candidates, anaplastic lymphoma kinase (ALK), was further investigated. Subsequent studies revealed that ALK promoted cell migration through the PI3K-AKT pathway via the p55γ regulatory subunit of PI3K, rather than more commonly used p85 subunit. Western blot and immunohistochemistry studies using mouse brain tissues revealed similar temporal expression patterns of ALK, phospho-p55γ, and phospho-AKT during different stages of development. These data support an important role for the p55γ subunit of PI3K in ALK-induced cell migration during brain development.
Keywords: ALK; PI3K; RNAi; brain development; cell migration.
Figures

Comment on
- Commentary to: RNAi-based functional selection identifies novel cell migration determinants dependent on PI3K and AKT pathways. Seo M, Lee S, Kim JH, Lee WH, Hu G, Elledge SJ, Suk K. Nat Commun 2014; 5:5217; PMID:25347953; https://doi.org/10.1038/ncomms6217
Similar articles
-
RNAi-based functional selection identifies novel cell migration determinants dependent on PI3K and AKT pathways.Nat Commun. 2014 Oct 28;5:5217. doi: 10.1038/ncomms6217. Nat Commun. 2014. PMID: 25347953 Free PMC article.
-
Activation of mammalian target of rapamycin signaling pathway contributes to tumor cell survival in anaplastic lymphoma kinase-positive anaplastic large cell lymphoma.Cancer Res. 2006 Jul 1;66(13):6589-97. doi: 10.1158/0008-5472.CAN-05-3018. Cancer Res. 2006. PMID: 16818631 Free PMC article.
-
Identification of PI3K regulatory subunit p55γ as a novel inhibitor of vascular smooth muscle cell proliferation and neointimal formation.Cardiovasc Res. 2015 Jan 1;105(1):75-85. doi: 10.1093/cvr/cvu235. Epub 2014 Nov 10. Cardiovasc Res. 2015. PMID: 25388664
-
Role of phosphatidylinositol 3-kinase-Akt pathway in nucleophosmin/anaplastic lymphoma kinase-mediated lymphomagenesis.Cancer Res. 2001 Mar 1;61(5):2194-9. Cancer Res. 2001. PMID: 11280786
-
Oncogenic tyrosine kinase NPM/ALK induces activation of the rapamycin-sensitive mTOR signaling pathway.Oncogene. 2007 Aug 16;26(38):5606-14. doi: 10.1038/sj.onc.1210346. Epub 2007 Mar 12. Oncogene. 2007. PMID: 17353907
Cited by
-
Defining Pathological Activities of ALK in Neuroblastoma, a Neural Crest-Derived Cancer.Int J Mol Sci. 2021 Oct 29;22(21):11718. doi: 10.3390/ijms222111718. Int J Mol Sci. 2021. PMID: 34769149 Free PMC article. Review.
-
TC2N: A Novel Vital Oncogene or Tumor Suppressor Gene In Cancers.Front Immunol. 2021 Dec 2;12:764749. doi: 10.3389/fimmu.2021.764749. eCollection 2021. Front Immunol. 2021. PMID: 34925334 Free PMC article. Review.
-
Identification of TC2N as a novel promising suppressor of PI3K-AKT signaling in breast cancer.Cell Death Dis. 2019 May 29;10(6):424. doi: 10.1038/s41419-019-1663-5. Cell Death Dis. 2019. PMID: 31142739 Free PMC article.
-
An analysis of the gene interaction networks identifying the role of PARP1 in metastasis of non-small cell lung cancer.Oncotarget. 2017 Aug 14;8(50):87263-87275. doi: 10.18632/oncotarget.20256. eCollection 2017 Oct 20. Oncotarget. 2017. PMID: 29152079 Free PMC article.
References
-
- Vicente-Manzanares M, Webb DJ, Horwitz AR. Cell migration at a glance. J Cell Sci 2005; 118:4917-9; PMID:16254237; https://doi.org/10.1242/jcs.02662 - DOI - PubMed
-
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science 2003; 302:1704-9; PMID:14657486; https://doi.org/10.1126/science.1092053 - DOI - PubMed
-
- Franz CM, Jones GE, Ridley AJ. Cell migration in development and disease. Dev Cell 2002; 2:153-8; PMID:11832241; https://doi.org/10.1016/S1534-5807(02)00120-X - DOI - PubMed
-
- Seo M, Lee S, Kim JH, Lee WH, Hu G, Elledge SJ, Suk K. RNAi-based functional selection identifies novel cell migration determinants dependent on PI3K and AKT pathways. Nat Commun 2014; 5:5217; PMID:25347953; https://doi.org/10.1038/ncomms6217 - DOI - PMC - PubMed
-
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al.. Cancer proliferation gene discovery through functional genomics. Science 2008; 319:620-4; PMID:18239126; https://doi.org/10.1126/science.1149200 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
