The Barley Powdery Mildew Effector Candidates CSEP0081 and CSEP0254 Promote Fungal Infection Success
- PMID: 27322386
- PMCID: PMC4913928
- DOI: 10.1371/journal.pone.0157586
The Barley Powdery Mildew Effector Candidates CSEP0081 and CSEP0254 Promote Fungal Infection Success
Abstract
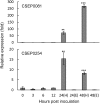
Effectors play significant roles in the success of pathogens. Recent advances in genome sequencing have revealed arrays of effectors and effector candidates from a wide range of plant pathogens. Yet, the vast majority of them remain uncharacterized. Among the ~500 Candidate Secreted Effector Proteins (CSEPs) predicted from the barley powdery mildew fungal genome, only a few have been studied and shown to have a function in virulence. Here, we provide evidence that CSEP0081 and CSEP0254 contribute to infection by the fungus. This was studied using Host-Induced Gene Silencing (HIGS), where independent silencing of the transcripts for these CSEPs significantly reduced the fungal penetration and haustoria formation rate. Both CSEPs are likely required during and after the formation of haustoria, in which their transcripts were found to be differentially expressed, rather than in epiphytic tissue. When expressed in barley leaf epidermal cells, both CSEPs appears to move freely between the cytosol and the nucleus, suggesting that their host targets locate in these cellular compartments. Collectively, our data suggest that, in addition to the previously reported effectors, the barley powdery mildew fungus utilizes these two CSEPs as virulence factors to enhance infection.
Conflict of interest statement
Figures

References
-
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444: 323–329. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

