Functional characterization of somatic mutations in cancer using network-based inference of protein activity
- PMID: 27322546
- PMCID: PMC5040167
- DOI: 10.1038/ng.3593
Functional characterization of somatic mutations in cancer using network-based inference of protein activity
Abstract
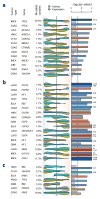
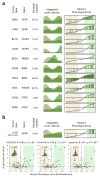
Identifying the multiple dysregulated oncoproteins that contribute to tumorigenesis in a given patient is crucial for developing personalized treatment plans. However, accurate inference of aberrant protein activity in biological samples is still challenging as genetic alterations are only partially predictive and direct measurements of protein activity are generally not feasible. To address this problem we introduce and experimentally validate a new algorithm, virtual inference of protein activity by enriched regulon analysis (VIPER), for accurate assessment of protein activity from gene expression data. We used VIPER to evaluate the functional relevance of genetic alterations in regulatory proteins across all samples in The Cancer Genome Atlas (TCGA). In addition to accurately infer aberrant protein activity induced by established mutations, we also identified a fraction of tumors with aberrant activity of druggable oncoproteins despite a lack of mutations, and vice versa. In vitro assays confirmed that VIPER-inferred protein activity outperformed mutational analysis in predicting sensitivity to targeted inhibitors.
Conflict of interest statement
MJA is chief scientific officer of DarwinHealth Inc. AC is a founder of DarwinHealth Inc.
Figures
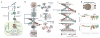

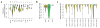
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

