Design and engineering of deimmunized biotherapeutics
- PMID: 27322891
- PMCID: PMC5067179
- DOI: 10.1016/j.sbi.2016.06.003
Design and engineering of deimmunized biotherapeutics
Abstract
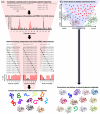
Therapeutic proteins are powerful next-generation drugs able to effectively treat diverse and devastating diseases, but the development and use of biotherapeutics entails unique challenges and risks. In particular, protein drugs are subject to immune surveillance in the human body, and ensuing antidrug immune responses can cause a wide range of problems including altered pharmacokinetics, loss of efficacy, and even life-threating complications. Here we review recent progress in technologies for engineering deimmunized biotherapeutics, placing particular emphasis on deletion of immunogenic antibody and T cell epitopes via experimentally or computationally guided mutagenesis.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
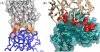
References
-
- Aggarwal S. What’s fueling the biotech engine - 2012 to 2013. Nat Biotech. 2014;32:32–39. - PubMed
-
- Schellekens H. The Immunogenicity of Therapeutic Proteins. Discovery Medicine. 2010;49:560–564. - PubMed
-
- Sethu S, Govindappa K, Alhaidari M, Pirmohamed M, Park K, Sathish J. Immunogenicity to Biologics: Mechanisms, Prediction and Reduction. Archivum Immunologiae Et Therapiae Experimentalis. 2012;60:331–344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

