Alveolar rhabdomyosarcoma: morphoproteomics and personalized tumor graft testing further define the biology of PAX3-FKHR(FOXO1) subtype and provide targeted therapeutic options
- PMID: 27323832
- PMCID: PMC5216796
- DOI: 10.18632/oncotarget.10089
Alveolar rhabdomyosarcoma: morphoproteomics and personalized tumor graft testing further define the biology of PAX3-FKHR(FOXO1) subtype and provide targeted therapeutic options
Abstract
Alveolar rhabdomyosarcoma (ARMS) represents a block in differentiation of malignant myoblasts. Genomic events implicated in the pathogenesis of ARMS involve PAX3-FKHR (FOXO1) or PAX7-FKHR (FOXO1) translocation with corresponding fusion transcripts and fusion proteins. Commonalities in ARMS include uncontrollable proliferation and failure to differentiate. The genomic-molecular correlates contributing to the etiopathogenesis of ARMS incorporate PAX3-FKHR (FOXO1) fusion protein stimulation of the IGF-1R, c-Met and GSK3-β pathways. With sequential morphoproteomic profiling on such a case in conjunction with personalized tumor graft testing, we provide an expanded definition of the biology of PAX3-FKHR (FOXO1) ARMS that integrates genomics, proteomics and pharmacogenomics. Moreover, therapies that target the genomic and molecular biology and lead to tumoral regression and/or tumoral growth inhibition in a xenograft model of ARMS are identified.
Significance: This case study could serve as a model for clinical trials using relatively low toxicity agents in both initial and maintenance therapies to induce remission and reduce the risk of recurrent disease in PAX3-FKHR (FOXO1) subtype of ARMS.
Keywords: PAX3-FKHR subtype; alveolar rhabdomyosarcoma; morphoproteomics; targeted therapy; xenograft testing.
Conflict of interest statement
The authors declared that they have no conflicts of interest to this work.
Figures


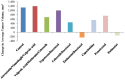
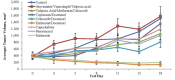
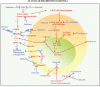
 ) and covered in detail in the Discussion. A proof of concept is provided in part by the results of xenograft testing of the patient's ARMS, PAX3-FKHR (FOXO1) subtype with the application of entinostat showing tumor regression and a combination of valproic acid, a class I histone deacetylase inhibitor, celecoxib and metformin showing tumor growth inhibition (see Figures 3 and 4, Table 1 and Discussion).
) and covered in detail in the Discussion. A proof of concept is provided in part by the results of xenograft testing of the patient's ARMS, PAX3-FKHR (FOXO1) subtype with the application of entinostat showing tumor regression and a combination of valproic acid, a class I histone deacetylase inhibitor, celecoxib and metformin showing tumor growth inhibition (see Figures 3 and 4, Table 1 and Discussion).References
-
- Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, Bridge JA, Crist WM, Triche TJ, Barr FG. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol. 2002;20:2672–2679. - PubMed
-
- Ayalon D, Glaser T, Werner H. Transcriptional regulation of IGF-I receptor gene expression by the PAX3-FKHR oncoprotein. Growth Horm IGF Res. 2001;11:289–297. - PubMed
-
- Ginsberg JP, Davis RJ, Bennicelli JL, Nauta LE, Barr FG. Up-regulation of MET but not neural cell adhesion molecule expression by the PAX3-FKHR fusion protein in alveolar rhabdomyosarcoma. Cancer Res. 1998;58:3542–3546. - PubMed
-
- Relaix F, Polimeni M, Rocancourt D, Ponzetto C, Schafer BW, Buckingham M. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes Dev. 2003;17:2950–2965. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

