Zebrafish cyclin Dx is required for development of motor neuron progenitors, and its expression is regulated by hypoxia-inducible factor 2α
- PMID: 27323909
- PMCID: PMC4915019
- DOI: 10.1038/srep28297
Zebrafish cyclin Dx is required for development of motor neuron progenitors, and its expression is regulated by hypoxia-inducible factor 2α
Abstract
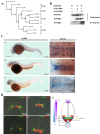
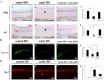
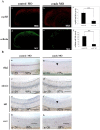
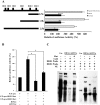
Cyclins play a central role in cell-cycle regulation; in mammals, the D family of cyclins consists of cyclin D1, D2, and D3. In Xenopus, only homologs of cyclins D1 and D2 have been reported, while a novel cyclin, cyclin Dx (ccndx), was found to be required for the maintenance of motor neuron progenitors during embryogenesis. It remains unknown whether zebrafish possess cyclin D3 or cyclin Dx. In this study, we identified a zebrafish ccndx gene encoding a protein which can form a complex with Cdk4. Through whole-mount in situ hybridization, we observed that zccndx mRNA is expressed in the motor neurons of hindbrain and spinal cord during development. Analysis of a 4-kb promoter sequence of the zccndx gene revealed the presence of HRE sites, which can be regulated by HIF2α. Morpholino knockdown of zebrafish Hif2α and cyclin Dx resulted in the abolishment of isl1 and oligo2 expression in the precursors of motor neurons, and also disrupted axon growth. Overexpression of cyclin Dx mRNA in Hif2α morphants partially rescued zccndx expression. Taken together, our data indicate that zebrafish cyclin Dx plays a role in maintaining the precursors of motor neurons.
Figures

References
-
- Hayles J. & Nurse P. Cell cycle regulation in yeast. J Cell Sci Suppl 4, 155–170 (1986). - PubMed
-
- Lee M. & Nurse P. Cell cycle control genes in fission yeast and mammalian cells. Trends Genet 4, 287–290(1988). - PubMed
-
- Pelech S. L., Sanghera J. S. & Daya-Makin M. Protein kinase cascades in meiotic and mitotic cell cycle control. Biochem Cell Biol 68, 1297–1330 (1990). - PubMed
-
- Maller J. L. Xenopus oocytes and the biochemistry of cell division. Biochemistry 29, 3157–3166 (1990). - PubMed
-
- Norbury C. & Nurse P. Animal cell cycles and their control. Annu Rev Biochem 61, 441–470 (1992). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

