The organization and regulation of mRNA-protein complexes
- PMID: 27324829
- PMCID: PMC5213448
- DOI: 10.1002/wrna.1369
The organization and regulation of mRNA-protein complexes
Abstract
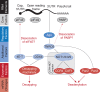
In a eukaryotic cell, each messenger RNA (mRNA) is bound to a variety of proteins to form an mRNA-protein complex (mRNP). Together, these proteins impact nearly every step in the life cycle of an mRNA and are critical for the proper control of gene expression. In the cytoplasm, for instance, mRNPs affect mRNA translatability and stability and provide regulation of specific transcripts as well as global, transcriptome-wide control. mRNPs are complex, diverse, and dynamic, and so they have been a challenge to understand. But the advent of high-throughput sequencing technology has heralded a new era in the study of mRNPs. Here, I will discuss general principles of cytoplasmic mRNP organization and regulation. Using microRNA-mediated repression as a case study, I will focus on common themes in mRNPs and highlight the interplay between mRNP composition and posttranscriptional regulation. mRNPs are an important control point in regulating gene expression, and while the study of these fascinating complexes presents remaining challenges, recent advances provide a critical lens for deciphering gene regulation. WIREs RNA 2017, 8:e1369. doi: 10.1002/wrna.1369 For further resources related to this article, please visit the WIREs website.
© 2016 The Authors. WIREs RNA published by Wiley Periodicals, Inc.
Figures



References
-
- Stevens A, Poole TL. 5′‐Exonuclease‐2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′‐exonuclease‐1. J Biol Chem 1995, 270:16063–16069. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

