The Zinc Finger of Prolyl Hydroxylase Domain Protein 2 Is Essential for Efficient Hydroxylation of Hypoxia-Inducible Factor α
- PMID: 27325674
- PMCID: PMC5007793
- DOI: 10.1128/MCB.00090-16
The Zinc Finger of Prolyl Hydroxylase Domain Protein 2 Is Essential for Efficient Hydroxylation of Hypoxia-Inducible Factor α
Abstract
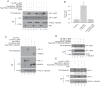
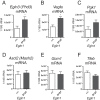
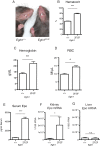
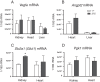
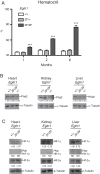
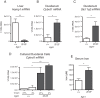
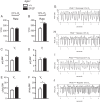
Prolyl hydroxylase domain protein 2 (PHD2) (also known as EGLN1) is a key oxygen sensor in mammals that posttranslationally modifies hypoxia-inducible factor α (HIF-α) and targets it for degradation. In addition to its catalytic domain, PHD2 contains an evolutionarily conserved zinc finger domain, which we have previously proposed recruits PHD2 to the HSP90 pathway to promote HIF-α hydroxylation. Here, we provide evidence that this recruitment is critical both in vitro and in vivo We show that in vitro, the zinc finger can function as an autonomous recruitment domain to facilitate interaction with HIF-α. In vivo, ablation of zinc finger function by a C36S/C42S Egln1 knock-in mutation results in upregulation of the erythropoietin gene, erythrocytosis, and augmented hypoxic ventilatory response, all hallmarks of Egln1 loss of function and HIF stabilization. Hence, the zinc finger ordinarily performs a critical positive regulatory function. Intriguingly, the function of this zinc finger is impaired in high-altitude-adapted Tibetans, suggesting that their adaptation to high altitude may, in part, be due to a loss-of-function EGLN1 allele. Thus, these findings have important implications for understanding both the molecular mechanism of the hypoxic response and human adaptation to high altitude.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim AA, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468–472. doi:10.1126/science.1059796. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
