Structure of a phleboviral envelope glycoprotein reveals a consolidated model of membrane fusion
- PMID: 27325770
- PMCID: PMC4932967
- DOI: 10.1073/pnas.1603827113
Structure of a phleboviral envelope glycoprotein reveals a consolidated model of membrane fusion
Abstract
An emergent viral pathogen termed severe fever with thrombocytopenia syndrome virus (SFTSV) is responsible for thousands of clinical cases and associated fatalities in China, Japan, and South Korea. Akin to other phleboviruses, SFTSV relies on a viral glycoprotein, Gc, to catalyze the merger of endosomal host and viral membranes during cell entry. Here, we describe the postfusion structure of SFTSV Gc, revealing that the molecular transformations the phleboviral Gc undergoes upon host cell entry are conserved with otherwise unrelated alpha- and flaviviruses. By comparison of SFTSV Gc with that of the prefusion structure of the related Rift Valley fever virus, we show that these changes involve refolding of the protein into a trimeric state. Reverse genetics and rescue of site-directed histidine mutants enabled localization of histidines likely to be important for triggering this pH-dependent process. These data provide structural and functional evidence that the mechanism of phlebovirus-host cell fusion is conserved among genetically and patho-physiologically distinct viral pathogens.
Keywords: bunyavirus; emerging virus; phlebovirus; structure; viral membrane fusion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
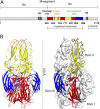






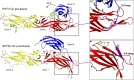
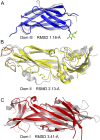
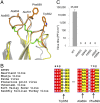
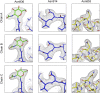
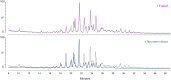

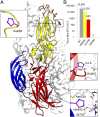
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

