GSK6853, a Chemical Probe for Inhibition of the BRPF1 Bromodomain
- PMID: 27326325
- PMCID: PMC4904261
- DOI: 10.1021/acsmedchemlett.6b00092
GSK6853, a Chemical Probe for Inhibition of the BRPF1 Bromodomain
Abstract
The BRPF (Bromodomain and PHD Finger-containing) protein family are important scaffolding proteins for assembly of MYST histone acetyltransferase complexes. A selective benzimidazolone BRPF1 inhibitor showing micromolar activity in a cellular target engagement assay was recently described. Herein, we report the optimization of this series leading to the identification of a superior BRPF1 inhibitor suitable for in vivo studies.
Keywords: BET; BRD1; BRPF1; BRPF2; BRPF3; bromodomain; chemical probe; epigenetics; inhibitor.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
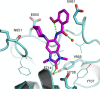
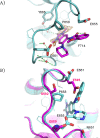


References
-
- Filippakopoulos P.; Picaud S.; Mangos M.; Keates T.; Lambert J. P.; Barsyte-Lovejoy D.; Felletar I.; Volkmer R.; Muller S.; Pawson T.; Gingras A. C.; Arrowsmith C. H.; Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. 10.1016/j.cell.2012.02.013. - DOI - PMC - PubMed
-
- Nicodeme E.; Jeffrey K. L.; Schaefer U.; Beinke S.; Dewell S.; Chung C. W.; Chandwani R.; Marazzi I.; Wilson P.; Coste H.; White J.; Kirilovsky J.; Rice C. M.; Lora J. M.; Prinjha R. K.; Lee K.; Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. 10.1038/nature09589. - DOI - PMC - PubMed
-
- Filippakopoulos P.; Qi J.; Picaud S.; Shen Y.; Smith W. B.; Fedorov O.; Morse E. M.; Keates T.; Hickman T. T.; Felletar I.; Philpott M.; Munro S.; McKeown M. R.; Wang Y.; Christie A. L.; West N.; Cameron M. J.; Schwartz B.; Heightman T. D.; La T. N.; French C. A.; Wiest O.; Kung A. L.; Knapp S.; Bradner J. E. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. 10.1038/nature09504. - DOI - PMC - PubMed
-
- Chung C.; Coste H.; White J. H.; Mirguet O.; Wilde J.; Gosmini R. L.; Delves C.; Magny S. M.; Woodward R.; Hughes S. A.; Boursier E. V.; Flynn H.; Bouillot A. M.; Bamborough P.; Brusq J. M.; Gellibert F. J.; Jones E. J.; Riou A. M.; Homes P.; Martin S. L.; Uings I. J.; Toum J.; Clement C. A.; Boullay A. B.; Grimley R. L.; Blandel F. M.; Prinjha R. K.; Lee K.; Kirilovsky J.; Nicodeme E. Discovery and Characterization of Small Molecule Inhibitors of the BET Family Bromodomains. J. Med. Chem. 2011, 54, 3827–3838. 10.1021/jm200108t. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

