Redesigning Aldolase Stereoselectivity by Homologous Grafting
- PMID: 27327271
- PMCID: PMC4915726
- DOI: 10.1371/journal.pone.0156525
Redesigning Aldolase Stereoselectivity by Homologous Grafting
Abstract
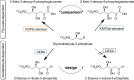
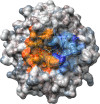
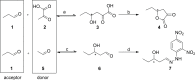
The 2-deoxy-d-ribose-5-phosphate aldolase (DERA) offers access to highly desirable building blocks for organic synthesis by catalyzing a stereoselective C-C bond formation between acetaldehyde and certain electrophilic aldehydes. DERA´s potential is particularly highlighted by the ability to catalyze sequential, highly enantioselective aldol reactions. However, its synthetic use is limited by the absence of an enantiocomplementary enzyme. Here, we introduce the concept of homologous grafting to identify stereoselectivity-determining amino acid positions in DERA. We identified such positions by structural analysis of the homologous aldolases 2-keto-3-deoxy-6-phosphogluconate aldolase (KDPG) and the enantiocomplementary enzyme 2-keto-3-deoxy-6-phosphogalactonate aldolase (KDPGal). Mutation of these positions led to a slightly inversed enantiopreference of both aldolases to the same extent. By transferring these sequence motifs onto DERA we achieved the intended change in enantioselectivity.
Conflict of interest statement
Figures


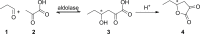
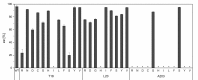
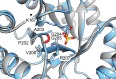
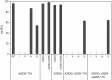
References
-
- Mugford PF, Wagner UG, Jiang Y, Faber K, Kazlauskas RJ. Enantiocomplementary Enzymes: Classification, Molecular Basis for Their Enantiopreference, and Prospects for Mirror‐Image Biotransformations. Angew Chem Int Ed. 2008; 47(46): 8782–93. - PubMed
-
- Gocke D, Walter L, Gauchenova E, Kolter G, Knoll M, Berthold C, et al. Rational Protein Design of ThDP-Dependent Enzymes—Engineering Stereoselectivity. Chem Express. 2008; 9(3): 406–12. - PubMed
-
- Hailes HC, Rother D, Müller M, Westphal R, Ward JM, Pleiss J, et al. Engineering stereoselectivity of ThDP-dependent enzymes. Exp Op Therap Patents. 2013; 280(24): 6374–94. - PubMed
-
- Pleiss J. Systematic Analysis of Large Enzyme Families: Identification of Specificity- and Selectivity-Determining Hotspots. ChemBioChem. 2014; 6(4): 944–50.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

