KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection
- PMID: 27327622
- PMCID: PMC4915687
- DOI: 10.1371/journal.ppat.1005700
KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection
Abstract
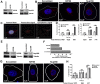
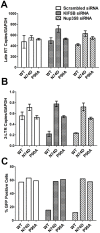
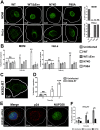
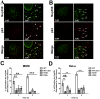
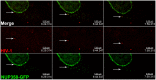
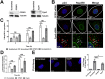
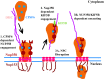
Following envelope mediated fusion, the HIV-1 core is released into the cytoplasm of the target cell and undergoes a series of trafficking and replicative steps that result in the nuclear import of the viral genome, which ultimately leads to the integration of the proviral DNA into the host cell genome. Previous studies have found that disruption of microtubules, or depletion of dynein or kinesin motors, perturb the normal uncoating and trafficking of the viral genome. Here, we show that the Kinesin-1 motor, KIF5B, induces a relocalization of the nuclear pore component Nup358 into the cytoplasm during HIV-1 infection. This relocalization of NUP358 is dependent on HIV-1 capsid, and NUP358 directly associates with viral cores following cytoplasmic translocation. This interaction between NUP358 and the HIV-1 core is dependent on multiple capsid binding surfaces, as this association is not observed following infection with capsid mutants in which a conserved hydrophobic binding pocket (N74D) or the cyclophilin A binding loop (P90A) is disrupted. KIF5B knockdown also prevents the nuclear entry and infection by HIV-1, but does not exert a similar effect on the N74D or P90A capsid mutants which do not rely on Nup358 for nuclear import. Finally, we observe that the relocalization of Nup358 in response to CA is dependent on cleavage protein and polyadenylation factor 6 (CPSF6), but independent of cyclophilin A. Collectively, these observations identify a previously unappreciated role for KIF5B in mediating the Nup358 dependent nuclear import of the viral genome during infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

