Structural insights into SAM domain-mediated tankyrase oligomerization
- PMID: 27328430
- PMCID: PMC5338228
- DOI: 10.1002/pro.2968
Structural insights into SAM domain-mediated tankyrase oligomerization
Abstract
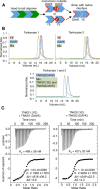
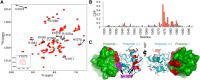
Tankyrase 1 (TNKS1; a.k.a. ARTD5) and tankyrase 2 (TNKS2; a.k.a ARTD6) are highly homologous poly(ADP-ribose) polymerases (PARPs) that function in a wide variety of cellular processes including Wnt signaling, Src signaling, Akt signaling, Glut4 vesicle translocation, telomere length regulation, and centriole and spindle pole maturation. Tankyrase proteins include a sterile alpha motif (SAM) domain that undergoes oligomerization in vitro and in vivo. However, the SAM domains of TNKS1 and TNKS2 have not been structurally characterized and the mode of oligomerization is not yet defined. Here we model the SAM domain-mediated oligomerization of tankyrase. The structural model, supported by mutagenesis and NMR analysis, demonstrates a helical, homotypic head-to-tail polymer that facilitates TNKS self-association. Furthermore, we show that TNKS1 and TNKS2 can form (TNKS1 SAM-TNKS2 SAM) hetero-oligomeric structures mediated by their SAM domains. Though wild-type tankyrase proteins have very low solubility, model-based mutations of the SAM oligomerization interface residues allowed us to obtain soluble TNKS proteins. These structural insights will be invaluable for the functional and biophysical characterization of TNKS1/2, including the role of TNKS oligomerization in protein poly(ADP-ribosyl)ation (PARylation) and PARylation-dependent ubiquitylation.
Keywords: PARP; PARP5; PARylation; SAM; molecular model; oligomerization; protein poly(ADP-ribosyl)ation; sterile alpha motif; tankyrase.
© 2016 The Protein Society.
Figures


References
-
- Huang S‐MA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461:614–620. - PubMed
-
- Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, Krzyzanowski PM, Scotter A, Gu S, Janmohamed S, Cong F, Simoncic PD, Ueki Y, La Rose J, Rottapel R (2011) Loss of Tankyrase‐mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell 147:1324–1339. - PMC - PubMed
-
- Smith S, De Lange T (2000) Tankyrase promotes telomere elongation in human cells. Curr Biol 10:1299–1302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

