The impact of genotype calling errors on family-based studies
- PMID: 27328765
- PMCID: PMC4916415
- DOI: 10.1038/srep28323
The impact of genotype calling errors on family-based studies
Abstract
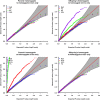
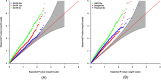
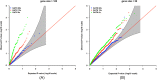
Family-based sequencing studies have unique advantages in enriching rare variants, controlling population stratification, and improving genotype calling. Standard genotype calling algorithms are less likely to call rare variants correctly, often mistakenly calling heterozygotes as reference homozygotes. The consequences of such non-random errors on association tests for rare variants are unclear, particularly in transmission-based tests. In this study, we investigated the impact of genotyping errors on rare variant association tests of family-based sequence data. We performed a comprehensive analysis to study how genotype calling errors affect type I error and statistical power of transmission-based association tests using a variety of realistic parameters in family-based sequencing studies. In simulation studies, we found that biased genotype calling errors yielded not only an inflation of type I error but also a power loss of association tests. We further confirmed our observation using exome sequence data from an autism project. We concluded that non-symmetric genotype calling errors need careful consideration in the analysis of family-based sequence data and we provided practical guidance on ameliorating the test bias.
Figures
References
-
- Pompanon F., Bonin A., Bellemain E. & Taberlet P. Genotyping errors: causes, consequences and solutions. Nature reviews. Genetics 6, 847–859 (2005). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

