Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer
- PMID: 27328849
- PMCID: PMC4917969
- DOI: 10.1038/ncomms12008
Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer
Abstract
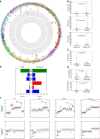
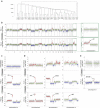
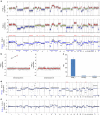
Genomic alterations in metastatic prostate cancer remain incompletely characterized. Here we analyse 493 prostate cancer cases from the TCGA database and perform whole-genome plasma sequencing on 95 plasma samples derived from 43 patients with metastatic prostate cancer. From these samples, we identify established driver aberrations in a cancer-related gene in nearly all cases (97.7%), including driver gene fusions (TMPRSS2:ERG), driver focal deletions (PTEN, RYBP and SHQ1) and driver amplifications (AR and MYC). In serial plasma analyses, we observe changes in focal amplifications in 40% of cases. The mean time interval between new amplifications was 26.4 weeks (range: 5-52 weeks), suggesting that they represent rapid adaptations to selection pressure. An increase in neuron-specific enolase is accompanied by clonal pattern changes in the tumour genome, most consistent with subclonal diversification of the tumour. Our findings suggest a high plasticity of prostate cancer genomes with newly occurring focal amplifications as a driving force in progression.
Figures

References
-
- Barbieri C. E. & Tomlins S. A. The prostate cancer genome: perspectives and potential. Urol. Oncol. 32, 53.e15–53.e22 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

