Degradation of the Plant Defense Signal Salicylic Acid Protects Ralstonia solanacearum from Toxicity and Enhances Virulence on Tobacco
- PMID: 27329752
- PMCID: PMC4916378
- DOI: 10.1128/mBio.00656-16
Degradation of the Plant Defense Signal Salicylic Acid Protects Ralstonia solanacearum from Toxicity and Enhances Virulence on Tobacco
Abstract
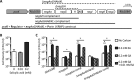
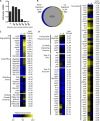
Plants use the signaling molecule salicylic acid (SA) to trigger defenses against diverse pathogens, including the bacterial wilt pathogen Ralstonia solanacearum SA can also inhibit microbial growth. Most sequenced strains of the heterogeneous R. solanacearum species complex can degrade SA via gentisic acid to pyruvate and fumarate. R. solanacearum strain GMI1000 expresses this SA degradation pathway during tomato pathogenesis. Transcriptional analysis revealed that subinhibitory SA levels induced expression of the SA degradation pathway, toxin efflux pumps, and some general stress responses. Interestingly, SA treatment repressed expression of virulence factors, including the type III secretion system, suggesting that this pathogen may suppress virulence functions when stressed. A GMI1000 mutant lacking SA degradation activity was much more susceptible to SA toxicity but retained the wild-type colonization ability and virulence on tomato. This may be because SA is less important than gentisic acid in tomato defense signaling. However, another host, tobacco, responds strongly to SA. To test the hypothesis that SA degradation contributes to virulence on tobacco, we measured the effect of adding this pathway to the tobacco-pathogenic R. solanacearum strain K60, which lacks SA degradation genes. Ectopic addition of the GMI1000 SA degradation locus, including adjacent genes encoding two porins and a LysR-type transcriptional regulator, significantly increased the virulence of strain K60 on tobacco. Together, these results suggest that R. solanacearum degrades plant SA to protect itself from inhibitory levels of this compound and also to enhance its virulence on plant hosts like tobacco that use SA as a defense signal molecule.
Importance: Plant pathogens such as the bacterial wilt agent Ralstonia solanacearum threaten food and economic security by causing significant losses for small- and large-scale growers of tomato, tobacco, banana, potato, and ornamentals. Like most plants, these crop hosts use salicylic acid (SA) both indirectly as a signal to activate defenses and directly as an antimicrobial chemical. We found that SA inhibits growth of R. solanacearum and induces a general stress response that includes repression of multiple bacterial wilt virulence factors. The ability to degrade SA reduces the pathogen's sensitivity to SA toxicity and increases its virulence on tobacco.
Copyright © 2016 Lowe-Power et al.
Figures


References
-
- Vidal S, Eriksson ARB, Montesano M, Denecke J, Palva ET. 1998. Cell wall degrading enzymes from Erwinia carotovora cooperate in the salicylic acid-independent induction of plant defense responses. Mol Plant Microbe Interact 11:23–32. doi: 10.1094/MPMI.1998.11.1.23. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
