Staphylococcus aureus Coordinates Leukocidin Expression and Pathogenesis by Sensing Metabolic Fluxes via RpiRc
- PMID: 27329753
- PMCID: PMC4916384
- DOI: 10.1128/mBio.00818-16
Staphylococcus aureus Coordinates Leukocidin Expression and Pathogenesis by Sensing Metabolic Fluxes via RpiRc
Abstract
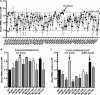
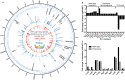
Staphylococcus aureus is a formidable human pathogen that uses secreted cytolytic factors to injure immune cells and promote infection of its host. Of these proteins, the bicomponent family of pore-forming leukocidins play critical roles in S. aureus pathogenesis. The regulatory mechanisms governing the expression of these toxins are incompletely defined. In this work, we performed a screen to identify transcriptional regulators involved in leukocidin expression in S. aureus strain USA300. We discovered that a metabolic sensor-regulator, RpiRc, is a potent and selective repressor of two leukocidins, LukED and LukSF-PV. Whole-genome transcriptomics, S. aureus exoprotein proteomics, and metabolomic analyses revealed that RpiRc influences the expression and production of disparate virulence factors. Additionally, RpiRc altered metabolic fluxes in the trichloroacetic acid cycle, glycolysis, and amino acid metabolism. Using mutational analyses, we confirmed and extended the observation that RpiRc signals through the accessory gene regulatory (Agr) quorum-sensing system in USA300. Specifically, RpiRc represses the rnaIII promoter, resulting in increased repressor of toxins (Rot) levels, which in turn negatively affect leukocidin expression. Inactivation of rpiRc phenocopied rot deletion and increased S. aureus killing of primary human polymorphonuclear leukocytes and the pathogenesis of bloodstream infection in vivo. Collectively, our results suggest that S. aureus senses metabolic shifts by RpiRc to differentially regulate the expression of leukocidins and to promote invasive disease.
Importance: The bicomponent pore-forming leukocidins play pivotal roles in the ability of S. aureus to kill multiple host immune cells, thus enabling this pathogen to have diverse tissue- and species-tropic effects. While the mechanisms of leukocidin-host receptor interactions have been studied in detail, the regulatory aspects of leukocidin expression are less well characterized. Moreover, the expression of the leukocidins is highly modular in vitro, suggesting the presence of regulators other than the known Agr, Rot, and S. aureus exoprotein pathways. Here, we describe how RpiRc, a metabolite-sensing transcription factor, mediates the repression of two specific leukocidin genes, lukED and pvl, which in turn has complex effects on the pathogenesis of S. aureus Our findings highlight the intricacies of leukocidin regulation by S. aureus and demonstrate the involvement of factors beyond traditional virulence factor regulators.
Copyright © 2016 Balasubramanian et al.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
