Has Rift Valley fever virus evolved with increasing severity in human populations in East Africa?
- PMID: 27329846
- PMCID: PMC4932650
- DOI: 10.1038/emi.2016.57
Has Rift Valley fever virus evolved with increasing severity in human populations in East Africa?
Abstract
Rift Valley fever (RVF) outbreaks have occurred across eastern Africa from 1912 to 2010 approximately every 4-15 years, most of which have not been accompanied by significant epidemics in human populations. However, human epidemics during RVF outbreaks in eastern Africa have involved 478 deaths in 1998, 1107 reported cases with 350 deaths from 2006 to 2007 and 1174 cases with 241 deaths in 2008. We review the history of RVF outbreaks in eastern Africa to identify the epidemiological factors that could have influenced its increasing severity in humans. Diverse ecological factors influence outbreak frequency, whereas virus evolution has a greater impact on its virulence in hosts. Several factors could have influenced the lack of information on RVF in humans during earlier outbreaks, but the explosive nature of human RVF epidemics in recent years mirrors the evolutionary trend of the virus. Comparisons between isolates from different outbreaks have revealed an accumulation of genetic mutations and genomic reassortments that have diversified RVF virus genomes over several decades. The threat to humans posed by the diversified RVF virus strains increases the potential public health and socioeconomic impacts of future outbreaks. Understanding the shifting RVF epidemiology as determined by its evolution is key to developing new strategies for outbreak mitigation and prevention of future human RVF casualties.
Figures

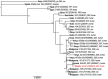
References
-
- Stordy RJ. Annual Report Department of Agriculture, British East Africa: 1912–1913. London: HMSO. 1913, 35.
-
- Daubney R, Hudson J, Garnham P. Enzootic hepatitis or Rift Valley fever. An undescribed virus disease of sheep cattle and man from East Africa. J Pathol Bacteriol 1931; 34: 545–579.
-
- Fyumagwa R, Ezekiel M, Nyaki A et al. Response to Rift Valley fever in Tanzania: challenges and opportunities. Tanzan J Health Res 2011; 13 (Suppl 1): 1–9. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
