Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals
- PMID: 27329977
- PMCID: PMC5854031
- DOI: 10.1093/molbev/msw112
Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals
Abstract
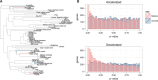
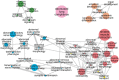
Mammal species have made the transition to the marine environment several times, and their lineages represent one of the classical examples of convergent evolution in morphological and physiological traits. Nevertheless, the genetic mechanisms of their phenotypic transition are poorly understood, and investigations into convergence at the molecular level have been inconclusive. While past studies have searched for convergent changes at specific amino acid sites, we propose an alternative strategy to identify those genes that experienced convergent changes in their selective pressures, visible as changes in evolutionary rate specifically in the marine lineages. We present evidence of widespread convergence at the gene level by identifying parallel shifts in evolutionary rate during three independent episodes of mammalian adaptation to the marine environment. Hundreds of genes accelerated their evolutionary rates in all three marine mammal lineages during their transition to aquatic life. These marine-accelerated genes are highly enriched for pathways that control recognized functional adaptations in marine mammals, including muscle physiology, lipid-metabolism, sensory systems, and skin and connective tissue. The accelerations resulted from both adaptive evolution as seen in skin and lung genes, and loss of function as in gustatory and olfactory genes. In regard to sensory systems, this finding provides further evidence that reduced senses of taste and smell are ubiquitous in marine mammals. Our analysis demonstrates the feasibility of identifying genes underlying convergent organism-level characteristics on a genome-wide scale and without prior knowledge of adaptations, and provides a powerful approach for investigating the physiological functions of mammalian genes.
Keywords: adaptive evolution; convergence; convergent evolution; functional constraint; marine mammals; relaxation of constraint.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Andersen HT. 1966. Physiological adaptations in diving vertebrates. Physiol Rev. 46:212–243. - PubMed
-
- Anisimova M, Bielawski JP, Yang Z. 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 18:1585–1592. - PubMed
-
- Arnason U, Gullberg A, Janke A, Kullberg M, Lehman N, Petrov EA, Väinölä R. 2006. Pinniped phylogeny and a new hypothesis for their origin and dispersal. Mol Phylogenet Evol. 41:345–354. - PubMed
-
- Bailey SF, Rodrigue N, Kassen R. 2015. The effect of selection environment on the probability of parallel evolution. Mol Biol Evol. 32:1436–1448. - PubMed
-
- Bills ML. 2011. Description of the chemical senses of the Florida manatee, Trichechus manatus latirostris, in relation to reproduction. [dissertation]. [Gainsville (FL)]: University of Florida.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

