Comparative analysis of EV isolation procedures for miRNAs detection in serum samples
- PMID: 27330048
- PMCID: PMC4916259
- DOI: 10.3402/jev.v5.31655
Comparative analysis of EV isolation procedures for miRNAs detection in serum samples
Abstract
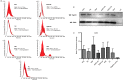
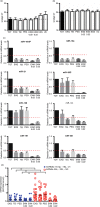
Extracellular vesicles (EVs) are emerging as potent non-invasive biomarkers. However, current methodologies are time consuming and difficult to translate to clinical practice. To analyse EV-encapsulated circulating miRNA, we searched for a quick, easy and economic method to enrich frozen human serum samples for EV. We compared the efficiency of several protocols and commercial kits to isolate EVs. Different methods based on precipitation, columns or filter systems were tested and compared with ultracentrifugation, which is the most classical protocol to isolate EVs. EV samples were assessed for purity and quantity by nanoparticle tracking analysis and western blot or cytometry against major EV protein markers. For biomarker validation, levels of a set of miRNAs were determined in EV fractions and compared with their levels in total serum. EVs isolated with precipitation-based methods were enriched for a subgroup of miRNAs that corresponded to miRNAs described to be encapsulated into EVs (miR-126, miR-30c and miR-143), while the detection of miR-21, miR-16-5p and miR-19a was very low compared with total serum. Our results point to precipitation using polyethylene glycol (PEG) as a suitable method for an easy and cheap enrichment of serum EVs for miRNA analyses. The overall performance of PEG was very similar, or better than other commercial precipitating reagents, in both protein and miRNA yield, but in comparison to them PEG is much cheaper. Other methods presented poorer results, mostly when assessing miRNA by qPCR analyses. Using PEG precipitation in a longitudinal study with human samples, we demonstrated that miRNA could be assessed in frozen samples up to 8 years of storage. We report a method based on a cut-off value of mean of fold EV detection versus serum that provides an estimate of the degree of encapsulation of a given miRNA.
Keywords: biomarker; diagnosis; extracellular vesicles; microRNA; polyethylene glycol; serum.
Figures



References
-
- Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer – new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21:470–9. - PubMed
-
- Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: association with disease and potential use as biomarkers. Crit Rev Oncol Hematol. 2011;80:193–208. - PubMed
-
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. - PubMed
-
- Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, et al. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977–83. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

