MET signaling in keratinocytes activates EGFR and initiates squamous carcinogenesis
- PMID: 27330189
- PMCID: PMC6322206
- DOI: 10.1126/scisignal.aaf5106
MET signaling in keratinocytes activates EGFR and initiates squamous carcinogenesis
Abstract
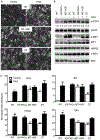
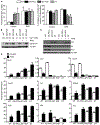
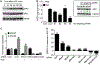
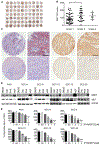
The receptor tyrosine kinase MET is abundant in many human squamous cell carcinomas (SCCs), but its functional significance in tumorigenesis is not clear. We found that the incidence of carcinogen-induced skin squamous tumors was substantially increased in transgenic MT-HGF (mouse metallothionein-hepatocyte growth factor) mice, which have increased abundance of the MET ligand HGF. Squamous tumors also erupted spontaneously on the skin of MT-HGF mice that were promoted by wounding or the application of 12-O-tetradecanoylphorbol 13-acetate, an activator of protein kinase C. Carcinogen-initiated tumors had Ras mutations, but spontaneous tumors did not. Cultured keratinocytes from MT-HGF mice and oncogenic RAS-transduced keratinocytes shared phenotypic and biochemical features of initiation that were dependent on autocrine activation of epidermal growth factor receptor (EGFR) through increased synthesis and release of EGFR ligands, which was mediated by the kinase SRC, the pseudoproteases iRhom1 and iRhom2, and the metallopeptidase ADAM17. Pharmacological inhibition of EGFR caused the regression of MT-HGF squamous tumors that developed spontaneously in orthografts of MT-HGF keratinocytes combined with dermal fibroblasts and implanted onto syngeneic mice. The global gene expression profile in MET-transformed keratinocytes was highly concordant with that in RAS-transformed keratinocytes, and a core RAS/MET coexpression network was activated in precancerous and cancerous human skin lesions. Tissue arrays revealed that many human skin SCCs have abundant HGF at both the transcript and protein levels. Thus, through the activation of EGFR, MET activation parallels a RAS pathway to contribute to human and mouse cutaneous cancers.
Copyright © 2016, American Association for the Advancement of Science.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures




References
-
- Mueller MM. Inflammation in epithelial skin tumours: old stories and new ideas. EurJCancer. 2006;42(6):735–44. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

