A Survey of Bioinformatics Database and Software Usage through Mining the Literature
- PMID: 27331905
- PMCID: PMC4917176
- DOI: 10.1371/journal.pone.0157989
A Survey of Bioinformatics Database and Software Usage through Mining the Literature
Abstract
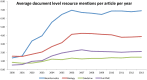
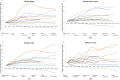
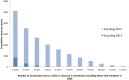
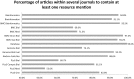
Computer-based resources are central to much, if not most, biological and medical research. However, while there is an ever expanding choice of bioinformatics resources to use, described within the biomedical literature, little work to date has provided an evaluation of the full range of availability or levels of usage of database and software resources. Here we use text mining to process the PubMed Central full-text corpus, identifying mentions of databases or software within the scientific literature. We provide an audit of the resources contained within the biomedical literature, and a comparison of their relative usage, both over time and between the sub-disciplines of bioinformatics, biology and medicine. We find that trends in resource usage differs between these domains. The bioinformatics literature emphasises novel resource development, while database and software usage within biology and medicine is more stable and conservative. Many resources are only mentioned in the bioinformatics literature, with a relatively small number making it out into general biology, and fewer still into the medical literature. In addition, many resources are seeing a steady decline in their usage (e.g., BLAST, SWISS-PROT), though some are instead seeing rapid growth (e.g., the GO, R). We find a striking imbalance in resource usage with the top 5% of resource names (133 names) accounting for 47% of total usage, and over 70% of resources extracted being only mentioned once each. While these results highlight the dynamic and creative nature of bioinformatics research they raise questions about software reuse, choice and the sharing of bioinformatics practice. Is it acceptable that so many resources are apparently never reused? Finally, our work is a step towards automated extraction of scientific method from text. We make the dataset generated by our study available under the CC0 license here: http://dx.doi.org/10.6084/m9.figshare.1281371.
Conflict of interest statement
Figures







References
-
- Gilbert D. Software review: Bioinformatics software resources. Briefings in Bioinformatics. 2004;5(3):300–304. - PubMed
-
- Babu PA, Udyama J, Kumar RK, Boddepalli R, Mangala DS, Rao GN. DoD2007: 1082 molecular biology databases. Bioinformation. 2007. January;2(2):64–7. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2174421/. 10.6026/97320630002064 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

