A Comparative Study of G-Quadruplex Unfolding and DNA Reeling Activities of Human RECQ5 Helicase
- PMID: 27332117
- PMCID: PMC4919422
- DOI: 10.1016/j.bpj.2016.05.016
A Comparative Study of G-Quadruplex Unfolding and DNA Reeling Activities of Human RECQ5 Helicase
Abstract
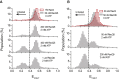
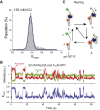
RECQ5 is one of five members of the RecQ family of helicases in humans, which include RECQ1, Bloom (BLM), Werner (WRN), RECQ4, and RECQ5. Both WRN and BLM have been shown to resolve G-quadruplex (GQ) structures. Deficiencies in unfolding GQ are known to result in DNA breaks and genomic instability, which are prominent in Werner and Bloom syndromes. RECQ5 is significant in suppressing sister chromatid exchanges during homologous recombination but its GQ unfolding activity are not known. We performed single-molecule studies under different salt (50-150 mM KCl or NaCl) and ATP concentrations on different GQ constructs including human telomeric GQ (with different overhangs and polarities) and GQ formed by thrombin-binding aptamer to investigate this activity. These studies demonstrated a RECQ5-mediated GQ unfolding activity that was an order of magnitude weaker than BLM and WRN. On the other hand, BLM and RECQ5 demonstrated similar single-stranded DNA (ssDNA) reeling activities that were not coupled to GQ unfolding. These results demonstrate overlap in function between these RecQ helicases; however, the relatively weak GQ destabilization activity of RECQ5 compared to BLM and WRN suggests that RECQ5 is not primarily associated with GQ destabilization, but could substitute for the more efficient helicases under conditions where their activity is compromised. In addition, these results implicate a more general role for helicase-promoted ssDNA reeling activity such as removal of proteins at the replication fork, whereas the association of ssDNA reeling with GQ destabilization is more helicase-specific.
Copyright © 2016 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Bachrati C.Z., Hickson I.D. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

