Escherichia coli Chromosomal Loci Segregate from Midcell with Universal Dynamics
- PMID: 27332118
- PMCID: PMC4919604
- DOI: 10.1016/j.bpj.2016.04.046
Escherichia coli Chromosomal Loci Segregate from Midcell with Universal Dynamics
Abstract
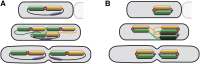
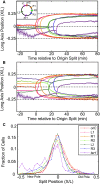
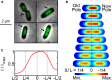
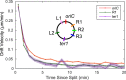
The structure of the Escherichia coli chromosome is inherently dynamic over the duration of the cell cycle. Genetic loci undergo both stochastic motion around their initial positions and directed motion to opposite poles of the rod-shaped cell during segregation. We developed a quantitative method to characterize cell-cycle dynamics of the E. coli chromosome to probe the chromosomal steady-state mobility and segregation process. By tracking fluorescently labeled chromosomal loci in thousands of cells throughout the entire cell cycle, our method allows for the statistical analysis of locus position and motion, the step-size distribution for movement during segregation, and the locus drift velocity. The robust statistics of our detailed analysis of the wild-type E. coli nucleoid allow us to observe loci moving toward midcell before segregation occurs, consistent with a replication factory model. Then, as segregation initiates, we perform a detailed characterization of the average segregation velocity of loci. Contrary to origin-centric models of segregation, which predict distinct dynamics for oriC-proximal versus oriC-distal loci, we find that the dynamics of loci were universal and independent of genetic position.
Copyright © 2016 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Cairns J. The bacterial chromosome and its manner of replication as seen by autoradiography. J. Mol. Biol. 1963;6:208–213. - PubMed
-
- Adachi S., Kohiyama M., Hiraga S. Localization of replication forks in wild-type and mukB mutant cells of Escherichia coli. Mol. Genet. Genomics. 2005;274:264–271. - PubMed
-
- den Blaauwen T., Aarsman M.E., Nanninga N. Pre-replication assembly of E. coli replisome components. Mol. Microbiol. 2006;62:695–708. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

