Using Clinical Trial Simulators to Analyse the Sources of Variance in Clinical Trials of Novel Therapies for Acute Viral Infections
- PMID: 27332704
- PMCID: PMC4917234
- DOI: 10.1371/journal.pone.0156622
Using Clinical Trial Simulators to Analyse the Sources of Variance in Clinical Trials of Novel Therapies for Acute Viral Infections
Abstract
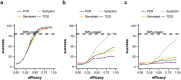
Background: About 90% of drugs fail in clinical development. The question is whether trials fail because of insufficient efficacy of the new treatment, or rather because of poor trial design that is unable to detect the true efficacy. The variance of the measured endpoints is a major, largely underestimated source of uncertainty in clinical trial design, particularly in acute viral infections. We use a clinical trial simulator to demonstrate how a thorough consideration of the variability inherent in clinical trials of novel therapies for acute viral infections can improve trial design.
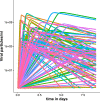
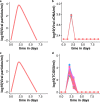
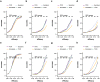
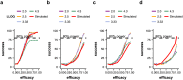
Methods and findings: We developed a clinical trial simulator to analyse the impact of three different types of variation on the outcome of a challenge study of influenza treatments for infected patients, including individual patient variability in the response to the drug, the variance of the measurement procedure, and the variance of the lower limit of quantification of endpoint measurements. In addition, we investigated the impact of protocol variation on clinical trial outcome. We found that the greatest source of variance was inter-individual variability in the natural course of infection. Running a larger phase II study can save up to $38 million, if an unlikely to succeed phase III trial is avoided. In addition, low-sensitivity viral load assays can lead to falsely negative trial outcomes.
Conclusions: Due to high inter-individual variability in natural infection, the most important variable in clinical trial design for challenge studies of potential novel influenza treatments is the number of participants. 100 participants are preferable over 50. Using more sensitive viral load assays increases the probability of a positive trial outcome, but may in some circumstances lead to false positive outcomes. Clinical trial simulations are powerful tools to identify the most important sources of variance in clinical trials and thereby help improve trial design.
Conflict of interest statement
Figures
References
-
- Mullard A. New drugs cost US$2.6 billion to develop. Nature Reviews Drug Discovery. 2014;13(12):877-.
-
- TCSDD TCftSoDD. Cost to Develop and Win Marketing Approval for a New Drug Is $2.6 Billion. 2014.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

