Revealing aperiodic aspects of solenoid proteins from sequence information
- PMID: 27334472
- PMCID: PMC6169467
- DOI: 10.1093/bioinformatics/btw319
Revealing aperiodic aspects of solenoid proteins from sequence information
Abstract
Motivation: Repeat proteins, which contain multiple repeats of short sequence motifs, form a large but seldom-studied group of proteins. Methods focusing on the analysis of 3D structures of such proteins identified many subtle effects in length distribution of individual motifs that are important for their functions. However, similar analysis was yet not applied to the vast majority of repeat proteins with unknown 3D structures, mostly because of the extreme diversity of the underlying motifs and the resulting difficulty to detect those.
Results: We developed FAIT, a sequence-based algorithm for the precise assignment of individual repeats in repeat proteins and introduced a framework to classify and compare aperiodicity patterns for large protein families. FAIT extracts repeat positions by post-processing FFAS alignment matrices with image processing methods. On examples of proteins with Leucine Rich Repeat (LRR) domains and other solenoids like proteins, we show that the automated analysis with FAIT correctly identifies exact lengths of individual repeats based entirely on sequence information.
Availability and implementation: https://github.com/GodzikLab/FAIT CONTACT: adam@godziklab.org
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
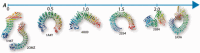


Similar articles
-
RAPHAEL: recognition, periodicity and insertion assignment of solenoid protein structures.Bioinformatics. 2012 Dec 15;28(24):3257-64. doi: 10.1093/bioinformatics/bts550. Epub 2012 Sep 8. Bioinformatics. 2012. PMID: 22962341
-
ConSole: using modularity of contact maps to locate solenoid domains in protein structures.BMC Bioinformatics. 2014 Apr 27;15:119. doi: 10.1186/1471-2105-15-119. BMC Bioinformatics. 2014. PMID: 24766872 Free PMC article.
-
Hammock: a hidden Markov model-based peptide clustering algorithm to identify protein-interaction consensus motifs in large datasets.Bioinformatics. 2016 Jan 1;32(1):9-16. doi: 10.1093/bioinformatics/btv522. Epub 2015 Sep 5. Bioinformatics. 2016. PMID: 26342231 Free PMC article.
-
Beta-rolls, beta-helices, and other beta-solenoid proteins.Adv Protein Chem. 2006;73:55-96. doi: 10.1016/S0065-3233(06)73003-0. Adv Protein Chem. 2006. PMID: 17190611 Review.
-
Comparison of ARM and HEAT protein repeats.J Mol Biol. 2001 May 25;309(1):1-18. doi: 10.1006/jmbi.2001.4624. J Mol Biol. 2001. PMID: 11491282 Review.
Cited by
-
Propagation of Fibrillar Structural Forms in Proteins Stopped by Naturally Occurring Short Polypeptide Chain Fragments.Pharmaceuticals (Basel). 2017 Nov 16;10(4):89. doi: 10.3390/ph10040089. Pharmaceuticals (Basel). 2017. PMID: 29144442 Free PMC article.
References
-
- Andrade M.A. et al. (2001) Protein repeats: structures, functions, and evolution. J. Struct. Biol., 134, 117–131. - PubMed
-
- Bazan J.F., Kajava A.V. (2015) Designs on a curve. Nat. Publ. Gr, 22, 103–105. - PubMed
-
- Biegert A., Söding J. (2008) De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics, 24, 807–814. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

