Crystal structure of raptor adenovirus 1 fibre head and role of the beta-hairpin in siadenovirus fibre head domains
- PMID: 27334597
- PMCID: PMC4918002
- DOI: 10.1186/s12985-016-0558-7
Crystal structure of raptor adenovirus 1 fibre head and role of the beta-hairpin in siadenovirus fibre head domains
Abstract
Background: Most adenoviruses recognize their host cells via an interaction of their fibre head domains with a primary receptor. The structural framework of adenovirus fibre heads is conserved between the different adenovirus genera for which crystal structures have been determined (Mastadenovirus, Aviadenovirus, Atadenovirus and Siadenovirus), but genus-specific differences have also been observed. The only known siadenovirus fibre head structure, that of turkey adenovirus 3 (TAdV-3), revealed a twisted beta-sandwich resembling the reovirus fibre head architecture more than that of other adenovirus fibre heads, plus a unique beta-hairpin embracing a neighbouring monomer. The TAdV-3 fibre head was shown to bind sialyllactose.
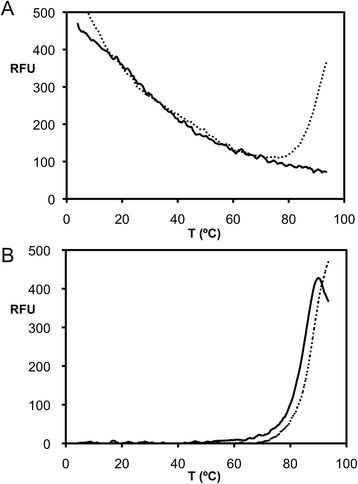
Methods: Raptor adenovirus 1 (RAdV-1) fibre head was expressed, crystallized and its structure was solved and refined at 1.5 Å resolution. The structure could be solved by molecular replacement using the TAdV-3 fibre head structure as a search model, despite them sharing a sequence identity of only 19 %. Versions of both the RAdV-1 and TAdV-3 fibre heads with their beta-hairpin arm deleted were prepared and their stabilities were compared with the non-mutated proteins by a thermal unfolding assay.
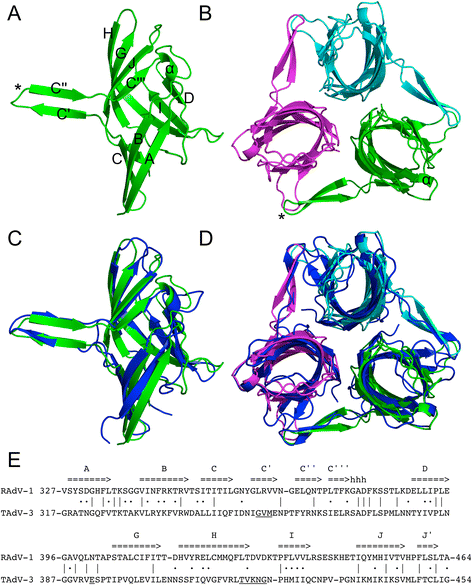
Results: The structure of the RAdV-1 fibre head contains the same twisted ABCJ-GHID beta-sandwich and beta-hairpin arm as the TAdV-3 fibre head. However, while the predicted electro-potential surface charge of the TAdV-3 fibre head is mainly positive, the RAdV-1 fibre head shows positively and negatively charged patches and does not appear to bind sialyllactose. Deletion of the beta-hairpin arm does not affect the structure of the raptor adenovirus 1 fibre head and only affects the stability of the RAdV-1 and TAdV-3 fibre heads slightly.
Conclusions: The high-resolution structure of RAdV-1 fibre head is the second known structure of a siadenovirus fibre head domain. The structure shows that the siadenovirus fibre head structure is conserved, but differences in the predicted surface charge suggest that RAdV-1 uses a different natural receptor for cell attachment than TAdV-3. Deletion of the beta-hairpin arm shows little impact on the structure and stability of the siadenovirus fibre heads.
Keywords: Atomic structure; Beta-hairpin; Deletion mutagenesis; Protein stability; Siadenovirus; X-ray crystallography.
Figures


References
-
- Wold WS, Horwitz MS. Adenoviruses. In: Fields BN, Knipe DM, Howley PM, editors. Fields virology. 5. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. pp. 2395–436.
-
- Benkő M. Adenoviruses: pathogenesis. In: Reference Module in Biomedical Sciences. Elsevier; 2015. doi:10.1016/B978-0-12-801238-3.02526-5.
-
- Waye MMY, Sing CW. Anti-viral drugs for human adenoviruses. Pharmaceuticals (Basel) 2010;3(10):3343–54. doi: 10.3390/ph3103343. - DOI
-
- Pihos AM. Epidemic keratoconjunctivitis: a review of current concepts in management. J Optom. 2013;6(2):69–74. doi: 10.1016/j.optom.2012.08.003. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

