Biological function of Foot-and-mouth disease virus non-structural proteins and non-coding elements
- PMID: 27334704
- PMCID: PMC4917953
- DOI: 10.1186/s12985-016-0561-z
Biological function of Foot-and-mouth disease virus non-structural proteins and non-coding elements
Abstract
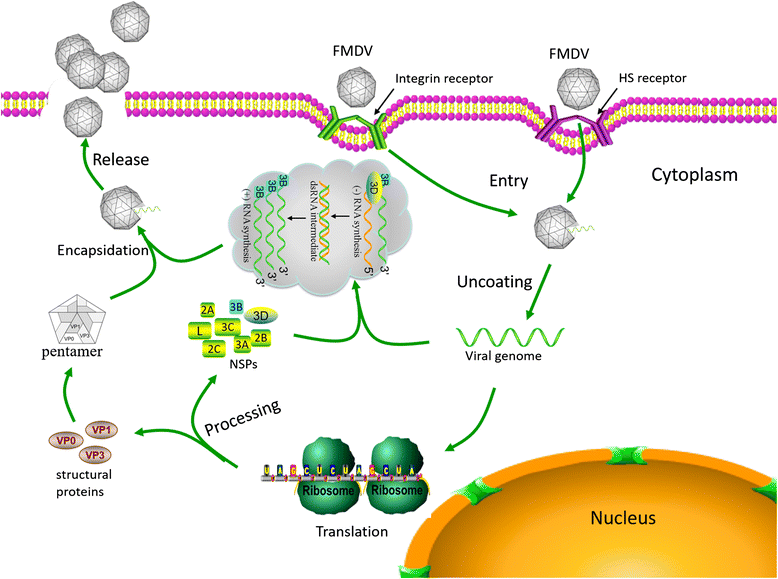
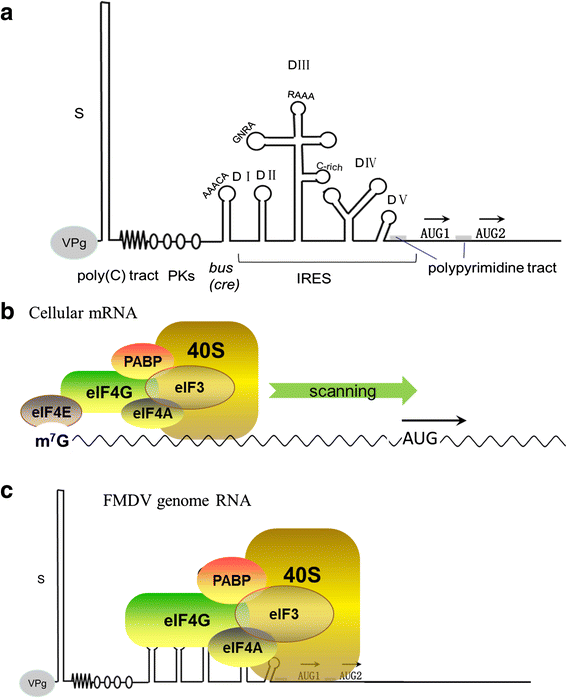
Foot-and-mouth disease virus (FMDV) represses host translation machinery, blocks protein secretion, and cleaves cellular proteins associated with signal transduction and the innate immune response to infection. Non-structural proteins (NSPs) and non-coding elements (NCEs) of FMDV play a critical role in these biological processes. The FMDV virion consists of capsid and nucleic acid. The virus genome is a positive single stranded RNA and encodes a single long open reading frame (ORF) flanked by a long structured 5'-untranslated region (5'-UTR) and a short 3'-UTR. The ORF is translated into a polypeptide chain and processed into four structural proteins (VP1, VP2, VP3, and VP4), 10 NSPs (L(pro), 2A, 2B, 2C, 3A, 3B1-3, 3C(pro), and 3D(pol)), and some cleavage intermediates. In the past decade, an increasing number of studies have begun to focus on the molecular pathogenesis of FMDV NSPs and NCEs. This review collected recent research progress on the biological functions of these NSPs and NCEs on the replication and host cellular regulation of FMDV to understand the molecular mechanism of host-FMDV interactions and provide perspectives for antiviral strategy and development of novel vaccines.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

