Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types
- PMID: 27335052
- PMCID: PMC5241901
- DOI: 10.1093/jnci/djw144
Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types
Abstract
Background: Immune infiltration of the tumor microenvironment has been associated with improved survival for some patients with solid tumors. The precise makeup and prognostic relevance of immune infiltrates across a broad spectrum of tumors remain unclear.
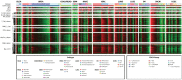
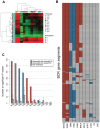
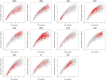
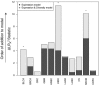
Methods: Using mRNA sequencing data from The Cancer Genome Atlas (TCGA) from 11 tumor types representing 3485 tumors, we evaluated lymphocyte and macrophage gene expression by tissue type and by genomic subtypes defined within and across tumor tissue of origin (Cox proportional hazards, Pearson correlation). We investigated clonal diversity of B-cell infiltrates through calculating B-cell receptor (BCR) repertoire sequence diversity. All statistical tests were two-sided.
Results: High expression of T-cell and B-cell signatures predicted improved overall survival across many tumor types including breast, lung, and melanoma (breast CD8_T_Cells hazard ratio [HR] = 0.36, 95% confidence interval [CI] = 0.16 to 0.81, P = .01; lung adenocarcinoma B_Cell_60gene HR = 0.71, 95% CI = 0.58 to 0.87, P = 7.80E-04; melanoma LCK HR = 0.86, 95% CI = 0.79 to 0.94, P = 6.75E-04). Macrophage signatures predicted worse survival in GBM, as did B-cell signatures in renal tumors (Glioblastoma Multiforme [GBM]: macrophages HR = 1.62, 95% CI = 1.17 to 2.26, P = .004; renal: B_Cell_60gene HR = 1.17, 95% CI = 1.04 to 1.32, P = .009). BCR diversity was associated with survival beyond gene segment expression in melanoma (HR = 2.67, 95% CI = 1.32 to 5.40, P = .02) and renal cell carcinoma (HR = 0.36, 95% CI = 0.15 to 0.87, P = .006).
Conclusions: These data support existing studies suggesting that in diverse tissue types, heterogeneous immune infiltrates are present and typically portend an improved prognosis. In some tumor types, BCR diversity was also associated with survival. Quantitative genomic signatures of immune cells warrant further testing as prognostic markers and potential biomarkers of response to cancer immunotherapy.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Balch CM, Riley LB, Bae YJ, et al. Patterns of human tumor-infiltrating lymphocytes in 120 human cancers. Arch Surg. 1990;125(2):200–205. - PubMed
-
- Kandalaft LE, Motz GT, Duraiswamy J, et al. Tumor immune surveillance and ovarian cancer: lessons on immune mediated tumor rejection or tolerance. Cancer Metastasis Rev. 2011;30(1):141–151. - PubMed
-
- Anagnostou VK, Brahmer JR. Cancer Immunotherapy: A Future Paradigm Shift in the Treatment of Non-Small Cell Lung Cancer. Clin Cancer Res. 2015;21(5):976–984. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

