The Evolution of Photoperiod-Insensitive Flowering in Sorghum, A Genomic Model for Panicoid Grasses
- PMID: 27335143
- PMCID: PMC4989116
- DOI: 10.1093/molbev/msw120
The Evolution of Photoperiod-Insensitive Flowering in Sorghum, A Genomic Model for Panicoid Grasses
Abstract
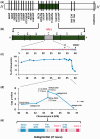
Of central importance in adapting plants of tropical origin to temperate cultivation has been selection of daylength-neutral genotypes that flower early in the temperate summer and take full advantage of its long days. A cross between tropical and temperate sorghums [Sorghum propinquum (Kunth) Hitchc.×S. bicolor (L.) Moench], revealed a quantitative trait locus (QTL), FlrAvgD1, accounting for 85.7% of variation in flowering time under long days. Fine-scale genetic mapping placed FlrAvgD1 on chromosome 6 within the physically largest centiMorgan in the genome. Forward genetic data from "converted" sorghums validated the QTL. Association genetic evidence from a diversity panel delineated the QTL to a 10-kb interval containing only one annotated gene, Sb06g012260, that was shown by reverse genetics to complement a recessive allele. Sb06g012260 (SbFT12) contains a phosphatidylethanolamine-binding (PEBP) protein domain characteristic of members of the "FT" family of flowering genes acting as a floral suppressor. Sb06g012260 appears to have evolved ∼40 Ma in a panicoid ancestor after divergence from oryzoid and pooid lineages. A species-specific Sb06g012260 mutation may have contributed to spread to temperate regions by S. halepense ("Johnsongrass"), one of the world's most widespread invasives. Alternative alleles for another family member, Sb02g029725 (SbFT6), mapping near another flowering QTL, also showed highly significant association with photoperiod response index (P = 1.53×10 (-) (6)). The evolution of Sb06g012260 adds to evidence that single gene duplicates play large roles in important environmental adaptations. Increased knowledge of Sb06g012260 opens new doors to improvement of sorghum and other grain and cellulosic biomass crops.
Keywords: FT domain; Sorghum halepense (“Johnsongrass”); conversion; flowering; photoperiod; single gene duplication..
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Allard R. 1956. Formulas and tables to facilitate the calculation of recombination values in heredity. Hilgardia 24:235–278.
-
- Berg JM, Tymoczko JL, Stryer L. 2002. Biochemistry. 5th ed New York: W.H: Freeman.
-
- Bowers JE, Abbey C, Anderson S, Chang C, Draye X, Hoppe AH, Jessup R, Lemke C, Lennington J, Li Z, et al. 2003. A high-density genetic recombination map of sequence-tagged sites for sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics 165:367–386. - PMC - PubMed
-
- Brady JA. 2006. Sorghum Ma5 and Ma6 maturity genes. College Station (TX: ): Texas A&M University.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

