The genetics of nodal marginal zone lymphoma
- PMID: 27335277
- PMCID: PMC5016706
- DOI: 10.1182/blood-2016-02-696757
The genetics of nodal marginal zone lymphoma
Abstract
Nodal marginal zone lymphoma (NMZL) is a rare, indolent B-cell tumor that is distinguished from splenic marginal zone lymphoma (SMZL) by the different pattern of dissemination. NMZL still lacks distinct markers and remains orphan of specific cancer gene lesions. By combining whole-exome sequencing, targeted sequencing of tumor-related genes, whole-transcriptome sequencing, and high-resolution single nucleotide polymorphism array analysis, we aimed at disclosing the pathways that are molecularly deregulated in NMZL and we compare the molecular profile of NMZL with that of SMZL. These analyses identified a distinctive pattern of nonsilent somatic lesions in NMZL. In 35 NMZL patients, 41 genes were found recurrently affected in ≥3 (9%) cases, including highly prevalent molecular lesions of MLL2 (also known as KMT2D; 34%), PTPRD (20%), NOTCH2 (20%), and KLF2 (17%). Mutations of PTPRD, a receptor-type protein tyrosine phosphatase regulating cell growth, were enriched in NMZL across mature B-cell tumors, functionally caused the loss of the phosphatase activity of PTPRD, and were associated with cell-cycle transcriptional program deregulation and increased proliferation index in NMZL. Although NMZL shared with SMZL a common mutation profile, NMZL harbored PTPRD lesions that were otherwise absent in SMZL. Collectively, these findings provide new insights into the genetics of NMZL, identify PTPRD lesions as a novel marker for this lymphoma across mature B-cell tumors, and support the distinction of NMZL as an independent clinicopathologic entity within the current lymphoma classification.
© 2016 by The American Society of Hematology.
Figures

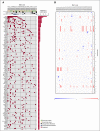

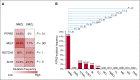


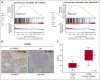
Comment in
-
Nodal marginal zone mutational signature.Blood. 2016 Sep 8;128(10):1315-6. doi: 10.1182/blood-2016-07-724963. Blood. 2016. PMID: 27609538 No abstract available.
References
-
- Swerdlow SH, Campo E, Harris NL, et al. Lyon, France: IARC; 2008. World Health Organization Classification of Tumours, Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues.
-
- Falini B, Agostinelli C, Bigerna B, et al. IRTA1 is selectively expressed in nodal and extranodal marginal zone lymphomas. Histopathology. 2012;61(5):930–941. - PubMed
-
- Falini B, Tiacci E, Pucciarini A, et al. Expression of the IRTA1 receptor identifies intraepithelial and subepithelial marginal zone B cells of the mucosa-associated lymphoid tissue (MALT). Blood. 2003;102(10):3684–3692. - PubMed
-
- Thieblemont C, Davi F, Noguera ME, Brière J. Non-MALT marginal zone lymphoma. Curr Opin Hematol. 2011;18(4):273–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

