Arf6 controls retromer traffic and intracellular cholesterol distribution via a phosphoinositide-based mechanism
- PMID: 27336679
- PMCID: PMC4931008
- DOI: 10.1038/ncomms11919
Arf6 controls retromer traffic and intracellular cholesterol distribution via a phosphoinositide-based mechanism
Abstract
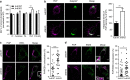
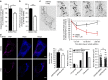
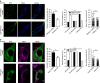
Small GTPases play a critical role in membrane traffic. Among them, Arf6 mediates transport to and from the plasma membrane, as well as phosphoinositide signalling and cholesterol homeostasis. Here we delineate the molecular basis for the link between Arf6 and cholesterol homeostasis using an inducible knockout (KO) model of mouse embryonic fibroblasts (MEFs). We find that accumulation of free cholesterol in the late endosomes/lysosomes of Arf6 KO MEFs results from mistrafficking of Niemann-Pick type C protein NPC2, a cargo of the cation-independent mannose-6-phosphate receptor (CI-M6PR). This is caused by a selective increase in an endosomal pool of phosphatidylinositol-4-phosphate (PI4P) and a perturbation of retromer, which controls the retrograde transport of CI-M6PR via sorting nexins, including the PI4P effector SNX6. Finally, reducing PI4P levels in KO MEFs through independent mechanisms rescues aberrant retromer tubulation and cholesterol mistrafficking. Our study highlights a phosphoinositide-based mechanism for control of cholesterol distribution via retromer.
Figures
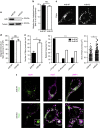
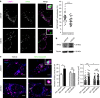
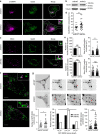
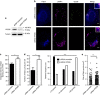

References
-
- Gillingham A. K. & Munro S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23, 579–611 (2007). - PubMed
-
- D'Souza-Schorey C., Li G., Colombo M. I. & Stahl P. D. A regulatory role for ARF6 in receptor-mediated endocytosis. Science 267, 1175–1178 (1995). - PubMed
-
- Donaldson J. G. Multiple roles for Arf6: sorting, structuring, and signalling at the plasma membrane. J. Biol. Chem. 278, 41573–41576 (2003). - PubMed
-
- D'Souza-Schorey C. & Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

