Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases
- PMID: 27337104
- PMCID: PMC4931022
- DOI: 10.1038/ncomms11974
Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases
Abstract
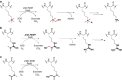
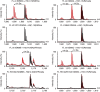
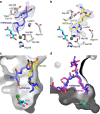
While the oxygen-dependent reversal of lysine N(ɛ)-methylation is well established, the existence of bona fide N(ω)-methylarginine demethylases (RDMs) is controversial. Lysine demethylation, as catalysed by two families of lysine demethylases (the flavin-dependent KDM1 enzymes and the 2-oxoglutarate- and oxygen-dependent JmjC KDMs, respectively), proceeds via oxidation of the N-methyl group, resulting in the release of formaldehyde. Here we report detailed biochemical studies clearly demonstrating that, in purified form, a subset of JmjC KDMs can also act as RDMs, both on histone and non-histone fragments, resulting in formaldehyde release. RDM catalysis is studied using peptides of wild-type sequences known to be arginine-methylated and sequences in which the KDM's methylated target lysine is substituted for a methylated arginine. Notably, the preferred sequence requirements for KDM and RDM activity vary even with the same JmjC enzymes. The demonstration of RDM activity by isolated JmjC enzymes will stimulate efforts to detect biologically relevant RDM activity.
Figures

References
-
- Walsh C. T., Garneau-Tsodikova S. & Gatto G. J. Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 44, 7342–7372 (2005). - PubMed
-
- Walport L. J., Hopkinson R. J. & Schofield C. J. Mechanisms of human histone and nucleic acid demethylases. Curr. Opin. Chem. Biol. 16, 525–534 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

