Programming a topologically constrained DNA nanostructure into a sensor
- PMID: 27337657
- PMCID: PMC4931013
- DOI: 10.1038/ncomms12074
Programming a topologically constrained DNA nanostructure into a sensor
Abstract
Many rationally engineered DNA nanostructures use mechanically interlocked topologies to connect individual DNA components, and their physical connectivity is achieved through the formation of a strong linking duplex. The existence of such a structural element also poses a significant topological constraint on functions of component rings. Herein, we hypothesize and confirm that DNA catenanes with a strong linking duplex prevent component rings from acting as the template for rolling circle amplification (RCA). However, by using an RNA-containing DNA [2] catenane with a strong linking duplex, we show that a stimuli-responsive RNA-cleaving DNAzyme can linearize one component ring, and thus enable RCA, producing an ultra-sensitive biosensing system. As an example, a DNA catenane biosensor is engineered to detect the model bacterial pathogen Escherichia coli through binding of a secreted protein, with a detection limit of 10 cells ml(-1), thus establishing a new platform for further applications of mechanically interlocked DNA nanostructures.
Figures
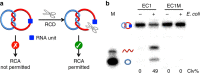



References
-
- Mao C., Sun W. & Seeman N. C. Assembly of Borromean rings from DNA. Nature 386, 137–138 (1997). - PubMed
-
- Wang H., Du S. M. & Seeman N. C. Tight single-stranded DNA knots. J. Biomol. Struct. Dyn. 10, 853–863 (1993). - PubMed
-
- Schmidt T. L. & Heckel A. Construction of a structurally defined double-stranded DNA catenane. Nano Lett. 11, 1739–1742 (2011). - PubMed
-
- Ackermann D. et al.. A double-stranded DNA rotaxane. Nat. Nanotechnol. 5, 436–442 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

