Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM)
- PMID: 27339025
- PMCID: PMC4919659
- DOI: 10.1038/srep28186
Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM)
Erratum in
-
Corrigendum: Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM).Sci Rep. 2016 Sep 22;6:33621. doi: 10.1038/srep33621. Sci Rep. 2016. PMID: 27654516 Free PMC article. No abstract available.
Abstract
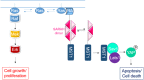
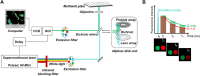
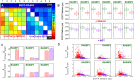
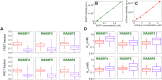
We present a high content multiwell plate cell-based assay approach to quantify protein interactions directly in cells using Förster resonance energy transfer (FRET) read out by automated fluorescence lifetime imaging (FLIM). Automated FLIM is implemented using wide-field time-gated detection, typically requiring only 10 s per field of view (FOV). Averaging over biological, thermal and shot noise with 100's to 1000's of FOV enables unbiased quantitative analysis with high statistical power. Plotting average donor lifetime vs. acceptor/donor intensity ratio clearly identifies protein interactions and fitting to double exponential donor decay models provides estimates of interacting population fractions that, with calibrated donor and acceptor fluorescence intensities, can yield dissociation constants. We demonstrate the application to identify binding partners of MST1 kinase and estimate interaction strength among the members of the RASSF protein family, which have important roles in apoptosis via the Hippo signalling pathway. KD values broadly agree with published biochemical measurements.
Figures






References
-
- Eggeling C., Willig K. I., Sahl S. J. & Hell S. W. Lens-based fluorescence nanoscopy. Quart. Rev. Biophys. 48, 178–243 (2015). - PubMed
-
- Zanella F., Lorens J. B. & Link W. High content screening: seeing is believing. Trends Biotechnol. 28, 237–245 (2010). - PubMed
-
- Jares-Erijman E. A. & Jovin T. M. Imaging molecular interactions in living cells by FRET microscopy. Curr. Opin. Chem. Biol. 10, 409–416 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

