GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes
- PMID: 27339137
- PMCID: PMC4948338
- DOI: 10.1073/pnas.1607769113
GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes
Abstract
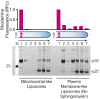
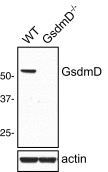
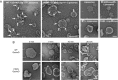
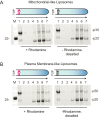
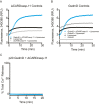
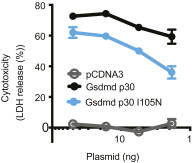
Gasdermin-D (GsdmD) is a critical mediator of innate immune defense because its cleavage by the inflammatory caspases 1, 4, 5, and 11 yields an N-terminal p30 fragment that induces pyroptosis, a death program important for the elimination of intracellular bacteria. Precisely how GsdmD p30 triggers pyroptosis has not been established. Here we show that human GsdmD p30 forms functional pores within membranes. When liberated from the corresponding C-terminal GsdmD p20 fragment in the presence of liposomes, GsdmD p30 localized to the lipid bilayer, whereas p20 remained in the aqueous environment. Within liposomes, p30 existed as higher-order oligomers and formed ring-like structures that were visualized by negative stain electron microscopy. These structures appeared within minutes of GsdmD cleavage and released Ca(2+) from preloaded liposomes. Consistent with GsdmD p30 favoring association with membranes, p30 was only detected in the membrane-containing fraction of immortalized macrophages after caspase-11 activation by lipopolysaccharide. We found that the mouse I105N/human I104N mutation, which has been shown to prevent macrophage pyroptosis, attenuated both cell killing by p30 in a 293T transient overexpression system and membrane permeabilization in vitro, suggesting that the mutants are actually hypomorphs, but must be above certain concentration to exhibit activity. Collectively, our data suggest that GsdmD p30 kills cells by forming pores that compromise the integrity of the cell membrane.
Keywords: GsdmD; caspase-11; pyroptosis.
Conflict of interest statement
Conflict of interest statement: R.A.A., A.E., A.G., P.S.L., N.K., C.C., V.M.D., and E.C.D. were employees of Genentech, Inc.
Figures






References
-
- Cookson BT, Brennan MA. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9(3):113–114. - PubMed
-
- Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–1022. - PubMed
-
- Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479(7371):117–121. - PubMed
-
- Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341(6151):1246–1249. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

