A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion
- PMID: 27339979
- PMCID: PMC5148821
- DOI: 10.1126/science.aaf7419
A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion
Abstract
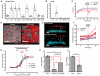
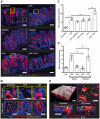
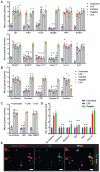
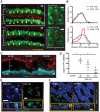
Innate immune signaling pathways contribute to the protection of host tissue when bacterially challenged. Colonic goblet cells are responsible for generating the two mucus layers that physically separate the luminal microbiota from the host epithelium. Analysis of colonic tissues from multiple mouse strains allowed us to identify a "sentinel" goblet cell (senGC) localized to the colonic crypt entrance. This cell nonspecifically endocytoses and reacts to the TLR2/1, TLR4, and TLR5 ligands by activating the Nlrp6 inflammasome downstream of TLR- and MyD88-dependent Nox/Duox reactive oxygen species synthesis. This triggers calcium ion-dependent compound exocytosis of Muc2 mucin from the senGC and generates an intercellular gap junction signal; in turn, this signal induces Muc2 secretion from adjacent goblet cells in the upper crypt, which expels bacteria. Thus, senGCs guard and protect the colonic crypt from bacterial intruders that have penetrated the inner mucus layer.
Copyright © 2016, American Association for the Advancement of Science.
Figures


Comment in
-
Intestinal tract: Sentinel goblet cells flush out bacteria from crypts.Nat Rev Gastroenterol Hepatol. 2016 Aug;13(8):438. doi: 10.1038/nrgastro.2016.117. Epub 2016 Jul 13. Nat Rev Gastroenterol Hepatol. 2016. PMID: 27407044 No abstract available.
-
Goblet cells as mucosal sentinels for immunity.Mucosal Immunol. 2017 Sep;10(5):1118-1121. doi: 10.1038/mi.2016.132. Epub 2017 Jan 25. Mucosal Immunol. 2017. PMID: 28120847 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

