Prevalence and diversity of H9N2 avian influenza in chickens of Northern Vietnam, 2014
- PMID: 27340015
- PMCID: PMC5036934
- DOI: 10.1016/j.meegid.2016.06.038
Prevalence and diversity of H9N2 avian influenza in chickens of Northern Vietnam, 2014
Abstract
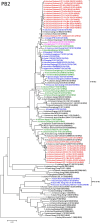
Despite their classification as low pathogenicity avian influenza viruses (LPAIV), A/H9N2 viruses cause significant losses in poultry in many countries throughout Asia, the Middle East and North Africa. To date, poultry surveillance in Vietnam has focused on detection of influenza H5 viruses, and there is limited understanding of influenza H9 epidemiology and transmission dynamics. We determined prevalence and diversity of influenza A viruses in chickens from live bird markets (LBM) of 7 northern Vietnamese provinces, using pooled oropharyngeal swabs collected from October to December 2014. Screening by real time RT-PCR revealed 1207/4900 (24.6%) of pooled swabs to be influenza A virus positive; overall prevalence estimates after accounting for pooling (5 swabs/pools) were 5.8% (CI 5.4-6.0). Subtyping was performed on 468 pooled swabs with M gene Ct<26. No influenza H7 was detected; 422 (90.1%) were H9 positive; and 22 (4.7%) were H5 positive. There was no evidence was of interaction between H9 and H5 virus detection rates. We sequenced 17 whole genomes of A/H9N2, 2 of A/H5N6, and 11 partial genomes. All H9N2 viruses had internal genes that clustered with genotype 57 and were closely related to Chinese human isolates of A/H7N9 and A/H10N8. Using a nucleotide divergence cutoff of 98%, we identified 9 distinct H9 genotypes. Phylogenetic analysis suggested multiple introductions of H9 viruses to northern Vietnam rather than in-situ transmission. Further investigations of H9 prevalence and diversity in other regions of Vietnam are warranted to assess H9 endemicity elsewhere in the country.
Keywords: Avian influenza; Chicken; H5; H9N2; Poultry; Vietnam; Zoonotic.
Copyright © 2016 The Authors. Published by Elsevier B.V. All rights reserved.
Figures








References
-
- Chen W.S., Calvo P.A., Malide D., Gibbs J., Schubert U., Bacik I.…Yewdell J.W. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001;7(12):1306–1312. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

