Mass spectrometry imaging for plant biology: a review
- PMID: 27340381
- PMCID: PMC4870303
- DOI: 10.1007/s11101-015-9440-2
Mass spectrometry imaging for plant biology: a review
Abstract
Mass spectrometry imaging (MSI) is a developing technique to measure the spatio-temporal distribution of many biomolecules in tissues. Over the preceding decade, MSI has been adopted by plant biologists and applied in a broad range of areas, including primary metabolism, natural products, plant defense, plant responses to abiotic and biotic stress, plant lipids and the developing field of spatial metabolomics. This review covers recent advances in plant-based MSI, general aspects of instrumentation, analytical approaches, sample preparation and the current trends in respective plant research.
Keywords: Biochemistry; Lateral resolution; Natural products; Spatial mapping; Spatial metabolomics.
Figures

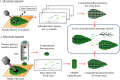

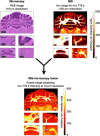


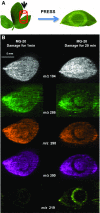
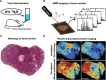
References
-
- Amstalden van Hove ER, Blackwell TR, Klinkert I, Eijkel GB, Heeren RMA, Glunde K. Multimodal mass spectrometric imaging of small molecules reveals distinct spatio-molecular signatures in differentially metastatic breast tumor models. Cancer Res. 2010;70:9012–9021. doi: 10.1158/0008-5472.CAN-10-0360. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
