Effective in silico prediction of new oxazolidinone antibiotics: force field simulations of the antibiotic-ribosome complex supervised by experiment and electronic structure methods
- PMID: 27340438
- PMCID: PMC4902031
- DOI: 10.3762/bjoc.12.45
Effective in silico prediction of new oxazolidinone antibiotics: force field simulations of the antibiotic-ribosome complex supervised by experiment and electronic structure methods
Erratum in
-
Correction: Effective in silico prediction of new oxazolidinone antibiotics: force field simulations of the antibiotic-ribosome complex supervised by experiment and electronic structure methods.Beilstein J Org Chem. 2016 Mar 31;12:608-10. doi: 10.3762/bjoc.12.59. eCollection 2016. Beilstein J Org Chem. 2016. PMID: 27346581 Free PMC article.
Abstract
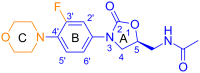
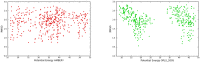
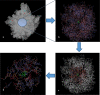
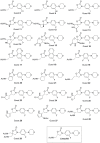
We propose several new and promising antibacterial agents for the treatment of serious Gram-positive infections. Our predictions rely on force field simulations, supervised by first principle calculations and available experimental data. Different force fields were tested in order to reproduce linezolid's conformational space in terms of a) the isolated and b) the ribosomal bound state. In a first step, an all-atom model of the bacterial ribosome consisting of nearly 1600 atoms was constructed and evaluated. The conformational space of 30 different ribosomal/oxazolidinone complexes was scanned by stochastic methods, followed by an evaluation of their enthalpic penalties or rewards and the mechanical strengths of the relevant hydrogen bonds (relaxed force constants; compliance constants). The protocol was able to reproduce the experimentally known enantioselectivity favoring the S-enantiomer. In a second step, the experimentally known MIC values of eight linezolid analogues were used in order to crosscheck the robustness of our model. In a final step, this benchmarking led to the prediction of several new and promising lead compounds. Synthesis and biological evaluation of the new compounds are on the way.
Keywords: compliance constants; computational chemistry; drug design; molecular recognition; relaxed force constants.
Figures



Similar articles
-
Correction: Effective in silico prediction of new oxazolidinone antibiotics: force field simulations of the antibiotic-ribosome complex supervised by experiment and electronic structure methods.Beilstein J Org Chem. 2016 Mar 31;12:608-10. doi: 10.3762/bjoc.12.59. eCollection 2016. Beilstein J Org Chem. 2016. PMID: 27346581 Free PMC article.
-
Synthesis and biological evaluation of novel 5-(hydroxamic acid)methyl oxazolidinone derivatives.Eur J Med Chem. 2015 Dec 1;106:120-31. doi: 10.1016/j.ejmech.2015.10.025. Epub 2015 Oct 23. Eur J Med Chem. 2015. PMID: 26536532
-
Linezolid, the first oxazolidinone antibacterial agent.Ann N Y Acad Sci. 2011 Mar;1222:49-54. doi: 10.1111/j.1749-6632.2011.05962.x. Ann N Y Acad Sci. 2011. PMID: 21434942 Review.
-
Tedizolid: a novel oxazolidinone with potent activity against multidrug-resistant gram-positive pathogens.Drugs. 2015 Feb;75(3):253-70. doi: 10.1007/s40265-015-0352-7. Drugs. 2015. PMID: 25673021 Review.
-
Treatment options for vancomycin-resistant enterococcal infections.Drugs. 2002;62(3):425-41. doi: 10.2165/00003495-200262030-00002. Drugs. 2002. PMID: 11827558 Review.
Cited by
-
Blinded evaluation of farnesoid X receptor (FXR) ligands binding using molecular docking and free energy calculations.J Comput Aided Mol Des. 2018 Jan;32(1):273-286. doi: 10.1007/s10822-017-0054-1. Epub 2017 Sep 2. J Comput Aided Mol Des. 2018. PMID: 28865056
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
