Marine-derived myxobacteria of the suborder Nannocystineae: An underexplored source of structurally intriguing and biologically active metabolites
- PMID: 27340488
- PMCID: PMC4902002
- DOI: 10.3762/bjoc.12.96
Marine-derived myxobacteria of the suborder Nannocystineae: An underexplored source of structurally intriguing and biologically active metabolites
Abstract
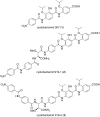
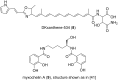
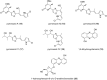
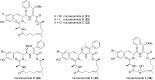
Myxobacteria are famous for their ability to produce most intriguing secondary metabolites. Till recently, only terrestrial myxobacteria were in the focus of research. In this review, however, we discuss marine-derived myxobacteria, which are particularly interesting due to their relatively recent discovery and due to the fact that their very existence was called into question. The to-date-explored members of these halophilic or halotolerant myxobacteria are all grouped into the suborder Nannocystineae. Few of them were chemically investigated revealing around 11 structural types belonging to the polyketide, non-ribosomal peptide, hybrids thereof or terpenoid class of secondary metabolites. A most unusual structural type is represented by salimabromide from Enhygromyxa salina. In silico analyses were carried out on the available genome sequences of four bacterial members of the Nannocystineae, revealing the biosynthetic potential of these bacteria.
Keywords: Enhygromyxa; Nannocystineae; genome mining; myxobacteria; natural products.
Figures









References
-
- Reichenbach H. Myxobacteria: A Most Peculiar Group of Social Prokaryotes. In: Rosenberg E, editor. Myxobacteria. New York, NY: Springer; 1984. pp. 1–50. ((Springer Series in Molecular Biology)). - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
