Nucleic Acid-Dependent Conformational Changes in CRISPR-Cas9 Revealed by Site-Directed Spin Labeling
- PMID: 27342128
- PMCID: PMC5183522
- DOI: 10.1007/s12013-016-0738-5
Nucleic Acid-Dependent Conformational Changes in CRISPR-Cas9 Revealed by Site-Directed Spin Labeling
Abstract
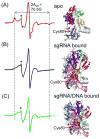
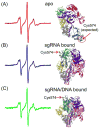
In a type II clustered regularly interspaced short palindromic repeats (CRISPR) system, RNAs that are encoded at the CRISPR locus complex with the CRISPR-associated (Cas) protein Cas9 to form an RNA-guided nuclease that cleaves double-stranded DNAs at specific sites. In recent years, the CRISPR-Cas9 system has been successfully adapted for genome engineering in a wide range of organisms. Studies have indicated that a series of conformational changes in Cas9, coordinated by the RNA and the target DNA, direct the protein into its active conformation, yet details on these conformational changes, as well as their roles in the mechanism of function of Cas9, remain to be elucidated. Here, nucleic acid-dependent conformational changes in Streptococcus pyogenes Cas9 (SpyCas9) were investigated using the method of site-directed spin labeling (SDSL). Single nitroxide spin labels were attached, one at a time, at one of the two native cysteine residues (Cys80 and Cys574) of SpyCas9, and the spin-labeled proteins were shown to maintain their function. X-band continuous-wave electron paramagnetic resonance spectra of the nitroxide attached at Cys80 revealed conformational changes of SpyCas9 that are consistent with a large-scale domain re-arrangement upon binding to its RNA partner. The results demonstrate the use of SDSL to monitor conformational changes in CRISPR-Cas9, which will provide key information for understanding the mechanism of CRISPR function.
Keywords: CRISPR; Cas9; Conformational change; EPR; Spin labeling.
Figures


Similar articles
-
Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9.Mol Cell. 2017 Jul 6;67(1):117-127.e5. doi: 10.1016/j.molcel.2017.05.024. Epub 2017 Jun 9. Mol Cell. 2017. PMID: 28602637 Free PMC article.
-
Probing the structural dynamics of the CRISPR-Cas9 RNA-guided DNA-cleavage system by coarse-grained modeling.Proteins. 2017 Feb;85(2):342-353. doi: 10.1002/prot.25229. Epub 2017 Jan 5. Proteins. 2017. PMID: 27936513
-
Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.PLoS Biol. 2019 Oct 11;17(10):e3000496. doi: 10.1371/journal.pbio.3000496. eCollection 2019 Oct. PLoS Biol. 2019. PMID: 31603896 Free PMC article.
-
[CRISPR/CAS9, the King of Genome Editing Tools].Mol Biol (Mosk). 2017 Jul-Aug;51(4):582-594. doi: 10.7868/S0026898417040036. Mol Biol (Mosk). 2017. PMID: 28900076 Review. Russian.
-
Optimization of genome editing through CRISPR-Cas9 engineering.Bioengineered. 2016 Apr;7(3):166-74. doi: 10.1080/21655979.2016.1189039. Bioengineered. 2016. PMID: 27340770 Free PMC article. Review.
Cited by
-
CRISPR-Cas9 Mediated DNA Unwinding Detected Using Site-Directed Spin Labeling.ACS Chem Biol. 2017 Jun 16;12(6):1489-1493. doi: 10.1021/acschembio.6b01137. Epub 2017 May 3. ACS Chem Biol. 2017. PMID: 28437608 Free PMC article.
-
Paramagnetic Chemical Probes for Studying Biological Macromolecules.Chem Rev. 2022 May 25;122(10):9571-9642. doi: 10.1021/acs.chemrev.1c00708. Epub 2022 Jan 27. Chem Rev. 2022. PMID: 35084831 Free PMC article. Review.
-
Kinetic Resolution of the Atomic 3D Structures Formed by Ground and Excited Conformational States in an RNA Dynamic Ensemble.J Am Chem Soc. 2023 Oct 25;145(42):22964-22978. doi: 10.1021/jacs.3c04614. Epub 2023 Oct 13. J Am Chem Soc. 2023. PMID: 37831584 Free PMC article.
-
Experimental Validation of the ALLNOX Program for Studying Protein-Nucleic Acid Complexes.J Phys Chem A. 2019 Apr 25;123(16):3592-3598. doi: 10.1021/acs.jpca.9b01027. Epub 2019 Apr 12. J Phys Chem A. 2019. PMID: 30978022 Free PMC article.
-
Structural integrity and side-chain interaction at the loop region of the bridge helix modulate Cas9 substrate discrimination.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf557. doi: 10.1093/nar/gkaf557. Nucleic Acids Res. 2025. PMID: 40613714 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

