EmpPrior: using outside empirical data to inform branch-length priors for Bayesian phylogenetics
- PMID: 27342194
- PMCID: PMC4919878
- DOI: 10.1186/s12859-016-1132-4
EmpPrior: using outside empirical data to inform branch-length priors for Bayesian phylogenetics
Abstract
Background: Branch-length parameters are a central component of phylogenetic models and of intrinsic biological interest. Default branch-length priors in some Bayesian phylogenetic software can be unintentionally informative and lead to branch- and tree-length estimates that are unreasonable. Alternatively, priors may be uninformative, but lead to diffuse posterior estimates. Despite the widespread availability of relevant datasets from other groups, biologists rarely leverage outside information to specify branch-length priors that are specific to the analysis they are conducting.
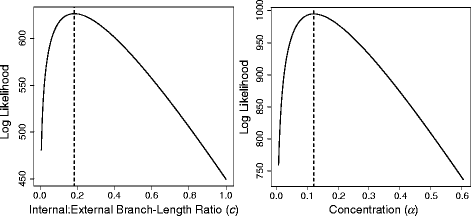
Results: We developed the software package EmpPrior to facilitate the collection and incorporation of relevant, outside information when setting branch-length priors for phylogenetics. EmpPrior efficiently queries TreeBASE to find data that are similar to focal data, in terms of taxonomic and genetic sampling, and uses them to inform branch-length priors for the focal analysis. EmpPrior consists of two components: EmpPrior-search, written in Java to query TreeBASE, and EmpPrior-fit, written in R to parameterize branch-length distributions. In an example analysis, we show how the use of relevant, outside data is made possible by EmpPrior and improves tree-length estimates from a focal dataset.
Conclusion: EmpPrior is easy to use, fast, and improves both the accuracy and precision of branch-length estimates in many circumstances. While EmpPrior's focus is on branch lengths, the strategy it employs could easily be extended to address other prior parameterization problems in phylogenetics.
Keywords: Bayesian phylogenetics; Branch lengths; Informed priors.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

