MicroRNA-1-associated effects of neuron-specific brain-derived neurotrophic factor gene deletion in dorsal root ganglia
- PMID: 27346077
- PMCID: PMC6675923
- DOI: 10.1016/j.mcn.2016.06.003
MicroRNA-1-associated effects of neuron-specific brain-derived neurotrophic factor gene deletion in dorsal root ganglia
Abstract
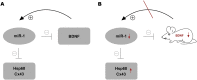
Background: MicroRNAs (miRNAs) regulate gene expression in physiological as well as in pathological processes, including chronic pain. Whether deletion of a gene can affect expression of the miRNAs that associate with the deleted gene mRNA remains elusive. We investigated the effects of brain-derived neurotrophic factor (Bdnf) gene deletion on the expression of miR-1 in dorsal root ganglion (DRG) neurons and its pain-associated downstream targets heat shock protein 60 (Hsp60) and connexin 43 (Cx43) in tamoxifen-inducible conditional knockout mice, Bdnf(fl/fl); Advillin-CreER(T2) (Bdnf cKO).
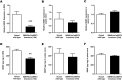
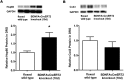
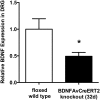
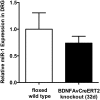
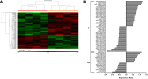
Results: Efficient Bdnf gene deletion was confirmed in DRG of Bdnf cKO mice by Real-Time qRT-PCR and ELISA 10days after completed tamoxifen treatment. In DRG, miR-1 expression was reduced 0.44-fold (p<0.05; Real-time qRT-PCR) in Bdnf cKO compared to floxed wildtype littermate control Bdnf(fl/fl) mice (WT). While Hsp60 protein expression was increased 1.85-fold (p<0.05; Western blot analysis), expression levels of Cx43 and the miR-1-associated transcription factors MEF2a and SRF remained unchanged. When analyzing Bdnf cKO mice 32days after complete tamoxifen treatment to investigate whether observed expression alterations remain permanently, we found no significant differences between Bdnf cKO and WT mice. However, miRNA microarray analysis revealed that 167 miRNAs altered (p<0.05) in DRG of these mice following Bdnf gene deletion.
Conclusions: Our results indicate that deletion of Bdnf in DRG neurons leads to a temporary dysregulation of miR-1, suggesting an impairment of a presumable feedback loop between BDNF protein and its targeting miR-1. This appears to affect its downstream protein Hsp60 and as a consequence might influence the phenotype after inducible Bdnf gene deletion. While this appears to be a MEF2a-/SRF-independent and transient effect, expression levels of various other miRNAs may remain permanently altered.
Keywords: BDNF-Advillin-Cre-ERT2; Bdnf; Dorsal root ganglion; Gene deletion; Neuropathic pain; miR-1; microRNA.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Expression changes of microRNA-1 and its targets Connexin 43 and brain-derived neurotrophic factor in the peripheral nervous system of chronic neuropathic rats.Mol Pain. 2015 Jun 26;11:39. doi: 10.1186/s12990-015-0045-y. Mol Pain. 2015. PMID: 26111928 Free PMC article.
-
Brain-derived neurotrophic factor increases in the uninjured dorsal root ganglion neurons in selective spinal nerve ligation model.J Neurosci. 2001 Jul 1;21(13):4891-900. doi: 10.1523/JNEUROSCI.21-13-04891.2001. J Neurosci. 2001. PMID: 11425916 Free PMC article.
-
Downregulation of miR-210 protected bupivacaine-induced neurotoxicity in dorsal root ganglion.Exp Brain Res. 2016 Apr;234(4):1057-65. doi: 10.1007/s00221-015-4513-4. Epub 2015 Dec 26. Exp Brain Res. 2016. PMID: 26708520
-
Conditional deletion of Pip5k1c in sensory ganglia and effects on nociception and inflammatory sensitization.Mol Pain. 2017 Jan-Dec;13:1744806917737907. doi: 10.1177/1744806917737907. Mol Pain. 2017. PMID: 29020859 Free PMC article.
-
The role of microRNAs in neurons and neuroimmune communication in the dorsal root ganglia in chronic pain.Neurosci Lett. 2020 Sep 14;735:135230. doi: 10.1016/j.neulet.2020.135230. Epub 2020 Jul 1. Neurosci Lett. 2020. PMID: 32621949 Review.
Cited by
-
Genes in Axonal Regeneration.Mol Neurobiol. 2024 Oct;61(10):7431-7447. doi: 10.1007/s12035-024-04049-z. Epub 2024 Feb 22. Mol Neurobiol. 2024. PMID: 38388774 Review.
-
MicroRNA-182 Alleviates Neuropathic Pain by Regulating Nav1.7 Following Spared Nerve Injury in Rats.Sci Rep. 2018 Nov 13;8(1):16750. doi: 10.1038/s41598-018-34755-3. Sci Rep. 2018. PMID: 30425258 Free PMC article.
-
Enhanced axonal response of mitochondria to demyelination offers neuroprotection: implications for multiple sclerosis.Acta Neuropathol. 2020 Aug;140(2):143-167. doi: 10.1007/s00401-020-02179-x. Epub 2020 Jun 22. Acta Neuropathol. 2020. PMID: 32572598 Free PMC article.
-
Role of Compensatory miRNA Networks in Cognitive Recovery from Heart Failure.Noncoding RNA. 2025 Jun 12;11(3):45. doi: 10.3390/ncrna11030045. Noncoding RNA. 2025. PMID: 40559623 Free PMC article.
-
Exosomal Chaperones and miRNAs in Gliomagenesis: State-of-Art and Theranostics Perspectives.Int J Mol Sci. 2018 Sep 5;19(9):2626. doi: 10.3390/ijms19092626. Int J Mol Sci. 2018. PMID: 30189598 Free PMC article. Review.
References
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bastian I., Tam Tam S., Zhou X.-F.F., Kazenwadel J., Van der Hoek M., Michael M.Z., Gibbins I., Haberberger R.V. Differential expression of microRNA-1 in dorsal root ganglion neurons. Histochem. Cell Biol. 2011;135:37–45. - PubMed
-
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
-
- Brandenburger T., Grievink H., Heinen N., Barthel F., Huhn R., Stachuletz F., Kohns M., Pannen B., Bauer I. Effects of remote ischemic preconditioning and myocardial ischemia on microRNA-1 expression in the rat heart in vivo. Shock. 2014;42:234–238. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

